Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells
- PMID: 11266458
- PMCID: PMC2195787
- DOI: 10.1083/jcb.152.4.657
Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells
Abstract
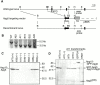
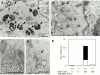
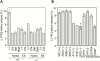
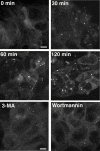
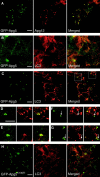
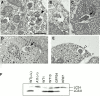
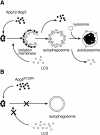
In macroautophagy, cytoplasmic components are delivered to lysosomes for degradation via autophagosomes that are formed by closure of cup-shaped isolation membranes. However, how the isolation membranes are formed is poorly understood. We recently found in yeast that a novel ubiquitin-like system, the Apg12-Apg5 conjugation system, is essential for autophagy. Here we show that mouse Apg12-Apg5 conjugate localizes to the isolation membranes in mouse embryonic stem cells. Using green fluorescent protein-tagged Apg5, we revealed that the cup-shaped isolation membrane is developed from a small crescent-shaped compartment. Apg5 localizes on the isolation membrane throughout its elongation process. To examine the role of Apg5, we generated Apg5-deficient embryonic stem cells, which showed defects in autophagosome formation. The covalent modification of Apg5 with Apg12 is not required for its membrane targeting, but is essential for involvement of Apg5 in elongation of the isolation membranes. We also show that Apg12-Apg5 is required for targeting of a mammalian Aut7/Apg8 homologue, LC3, to the isolation membranes. These results suggest that the Apg12-Apg5 conjugate plays essential roles in isolation membrane development.
Figures


References
-
- Baba M., Ohusumi M., Ohsumi Y. Analysis of the membrane structure involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. - PubMed
-
- Clarke P.G. Developmental cell deathmorphological diversity and multiple mechanisms. Anat. Embryol. 1990;181:195–213. - PubMed
-
- Dice J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biol. Sci. 1990;15:305–309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

