All Tcf HMG box transcription factors interact with Groucho-related co-repressors
- PMID: 11266540
- PMCID: PMC31284
- DOI: 10.1093/nar/29.7.1410
All Tcf HMG box transcription factors interact with Groucho-related co-repressors
Abstract
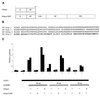
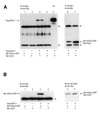
Tcf/Lef family transcription factors are the downstream effectors of the Wingless/Wnt signal transduction pathway. Upon Wingless/Wnt signalling, beta-catenin translocates to the nucleus, interacts with Tcf (1-3) and thus activates transcription of target genes (4,5). Tcf factors also interact with members of the Groucho (Grg/TLE) family of transcriptional co-repressors (6). We have now tested all known mammalian Groucho family members for their ability to interact specifically with individual Tcf/Lef family members. Transcriptional activation by any Tcf could be repressed by Grg-1, Grg-2/TLE-2, Grg-3 and Grg-4 in a reporter assay. Specific interactions between Tcf and Grg proteins may be achieved in vivo by tissue- or cell type-limited expression. To address this, we determined the expression of all Tcf and Grg/TLE family members in a panel of cell lines. Within any cell line, several Tcfs and TLEs are co-expressed. Thus, redundancy in Tcf/Grg interactions appears to be the rule. The 'long' Groucho family members containing five domains are repressors of Tcf-mediated transactivation, whereas Grg-5, which only contains the first two domains, acts as a de-repressor. As previously shown for Drosophila Groucho, we show that long Grg proteins interact with histone deacetylase-1. Although Grg-5 contains the GP homology domain that mediates HDAC binding in long Grg proteins, Grg-5 fails to bind this co-repressor, explaining how it can de-repress transcription.
Figures
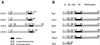






References
-
- van de Wetering M., Cavallo,R., Dooijes,D., van Beest,M., van Es,J., Loureiro,J., Ypma,A., Hursh,D., Jones,T., Bejsovec,A., Peifer,M., Mortin,M. and Clevers,H. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. - PubMed
-
- Molenaar M., van de Wetering,M., Oosterwegel,M., Peterson Maduro,J., Godsave,S., Korinek,V., Roose,J., Destree,O. and Clevers,H. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell, 86, 391–399. - PubMed
-
- Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. - PubMed
-
- He T.C., Sparks,A.B., Rago,C., Hermeking,H., Zawel,L., da Costa,L.T., Morin,P.J., Vogelstein,B. and Kinzler,K.W. (1998) Identification of c-MYC as a target of the APC pathway. Science, 281, 1509–1512. - PubMed
-
- Tetsu O. and McCormick,F. (1999) β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

