DNA methyltransferases of the cyanobacterium Anabaena PCC 7120
- PMID: 11266551
- PMCID: PMC31280
- DOI: 10.1093/nar/29.7.1491
DNA methyltransferases of the cyanobacterium Anabaena PCC 7120
Abstract
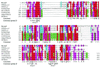
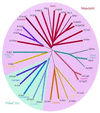
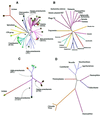
From the characterization of enzyme activities and the analysis of genomic sequences, the complement of DNA methyltransferases (MTases) possessed by the cyanobacterium ANABAENA PCC 7120 has been deduced. ANABAENA has nine DNA MTases. Four are associated with Type II restriction enzymes (AVAI, AVAII, AVAIII and the newly recognized inactive AVAIV), and five are not. Of the latter, four may be classified as solitary MTases, those whose function lies outside of a restriction/modification system. The group is defined here based on biochemical and genetic characteristics. The four solitary MTases, DmtA/M.AVAVI, DmtB/M.AVAVII, DmtC/M. AVAVIII and DmtD/M.AVAIX, methylate at GATC, GGCC, CGATCG and rCCGGy, respectively. DmtB methylates cytosines at the N4 position, but its sequence is more similar to N6-adenine MTases than to cytosine-specific enzymes, indicating that it may have evolved from the former. The solitary MTases, appear to be of ancient origin within cyanobacteria, while the restriction MTases appear to have arrived by recent horizontal transfer as did five now inactive Type I restriction systems. One Mtase, M.AVAV, cannot reliably be classified as either a solitary or restriction MTase. It is structurally unusual and along with a few proteins of prokaryotic and eukaryotic origin defines a structural class of MTases distinct from all previously described.
Figures


References
-
- Wilson G.G. and Murray,N.E. (1991) Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. - PubMed
-
- Palmer B.R. and Marinus,M.G. (1994) The dam and dcm strains of Escherichia coli – a review. Gene, 143, 1–12. - PubMed
-
- Zweiger G., Marcynski,G. and Shapiro,L. (1994) A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol., 235, 472–485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

