A single alteration 20 nt 5' to an editing target inhibits chloroplast RNA editing in vivo
- PMID: 11266552
- PMCID: PMC31290
- DOI: 10.1093/nar/29.7.1507
A single alteration 20 nt 5' to an editing target inhibits chloroplast RNA editing in vivo
Abstract
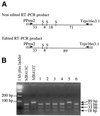
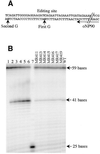
Transcripts of typical dicot plant plastid genes undergo C-->U RNA editing at approximately 30 locations, but there is no consensus sequence surrounding the C targets of editing. The cis-acting elements required for editing of the C located at tobacco rpoB editing site II were investigated by introducing translatable chimeric minigenes containing sequence -20 to +6 surrounding the C target of editing. When the -20 to +6 sequence specified by the homologous region present in the black pine chloroplast genome was incorporated, virtually no editing of the transcripts occurred in transgenic tobacco plastids. Nucleotides that differ between the black pine and tobacco sequence were tested for their role in C-->U editing by designing chimeric genes containing one or more of these divergent nucleotides. Surprisingly, the divergent nucleotide that had the strongest negative effect on editing of the minigene transcript was located -20 nt 5' to the C target of editing. Expression of transgene transcripts carrying the 27 nt sequence did not affect the editing extent of the endogenous rpoB transcripts, even though the chimeric transcripts were much more abundant than those of the endogenous gene. In plants carrying a 93 nt rpoB editing site sequence, transgene transcripts accumulated to a level three times greater than transgene transcripts in the plants carrying the 27 nt rpoB editing sites and resulted in editing of the endogenous transcripts from 100 to 50%. Both a lower affinity of the 27 nt site for a trans-acting factor and lower abundance of the transcript could explain why expression of minigene transcripts containing the 27 nt sequence did not affect endogenous editing.
Figures



References
-
- Maier R.M., Neckermann,K., Igloi,G.L. and Kössel,H. (1995) Complete sequence of the maize chloroplast genome: gene content, hot-spots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol., 251, 614–628. - PubMed
-
- Zito F, Kuras,R., Choquet,Y., Kossel,H. and Wollman,F.A. (1997) Mutations of cytochrome b(6) in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol., 33, 79–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

