Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms
- PMID: 11266558
- PMCID: PMC31286
- DOI: 10.1093/nar/29.7.1556
Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms
Abstract
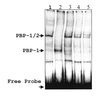
Typical general transcription factors, such as TATA binding protein and TFII B, have not yet been identified in any member of the Trypanosomatidae family of parasitic protozoa. Interestingly, mRNA coding genes do not appear to have discrete transcriptional start sites, although in most cases they require an RNA polymerase that has the biochemical properties of eukaryotic RNA polymerase II. A discrete transcription initiation site may not be necessary for mRNA synthesis since the sequences upstream of each transcribed coding region are trimmed from the nascent transcript when a short m(7)G-capped RNA is added during mRNA maturation. This short 39 nt m(7)G-capped RNA, the spliced leader (SL) sequence, is expressed as an approximately 100 nt long RNA from a set of reiterated, though independently transcribed, genes in the trypanosome genome. Punctuation of the 5' end of mRNAs by a m(7)G cap-containing spliced leader is a developing theme in the lower eukaryotic world; organisms as diverse as EUGLENA: and nematode worms, including Caenorhabditis elegans, utilize SL RNA in their mRNA maturation programs. Towards understanding the coordination of SL RNA and mRNA expression in trypanosomes, we have begun by characterizing SL RNA gene expression in the model trypanosome Leptomonas seymouri. Using a homologous in vitro transcription system, we demonstrate in this study that the SL RNA is transcribed by RNA polymerase II. During SL RNA transcription, accurate initiation is determined by an initiator element with a loose consensus of CYAC/AYR(+1). This element, as well as two additional basal promoter elements, is divergent in sequence from the basal transcription elements seen in other eukaryotic gene promoters. We show here that the in vitro transcription extract contains a binding activity that is specific for the initiator element and thus may participate in recruiting RNA polymerase II to the SL RNA gene promoter.
Figures




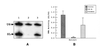

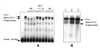

References
-
- Bangs J.D., Crain,P.F., Hashizume,T., McCloskey,J.A. and Boothroyd,J.C. (1992) Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem., 267, 9805–9815. - PubMed
-
- Günzl A., Ullu,E., Dörner,M., Fragoso,S., Hoffmann,K., Milner,J., Morita,Y., Nguu,E., Vanacova,S., Wünsch,S., Dare,A., Kwon,H. and Tschudi,C. (1997) Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol., 85, 67–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

