Identification of thermophilic species by the amino acid compositions deduced from their genomes
- PMID: 11266564
- PMCID: PMC31282
- DOI: 10.1093/nar/29.7.1608
Identification of thermophilic species by the amino acid compositions deduced from their genomes
Abstract
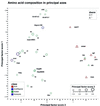
The global amino acid compositions as deduced from the complete genomic sequences of six thermophilic archaea, two thermophilic bacteria, 17 mesophilic bacteria and two eukaryotic species were analysed by hierarchical clustering and principal components analysis. Both methods showed an influence of several factors on amino acid composition. Although GC content has a dominant effect, thermophilic species can be identified by their global amino acid compositions alone. This study presents a careful statistical analysis of factors that affect amino acid composition and also yielded specific features of the average amino acid composition of thermophilic species. Moreover, we introduce the first example of a 'compositional tree' of species that takes into account not only homologous proteins, but also proteins unique to particular species. We expect this simple yet novel approach to be a useful additional tool for the study of phylogeny at the genome level.
Figures



References
-
- McDonald J.H., Grasso,A.M. and Rejto,L.K. (1999) Patterns of temperature adaption in proteins from Methanococcus and Bacillus. Mol. Biol. Evol., 16, 1785–1790. - PubMed
-
- Jaenicke R. and Böhm,G. (1998) The stability of proteins in extreme environments. Curr. Opin. Struct. Biol., 8, 738–748. - PubMed
-
- Perutz M.F. (1978) Electrostatic effects in proteins. Science, 201, 1187–1191. - PubMed
-
- Kreil D.P. and Etzold,T.M. (2000) SRS—access to molecular biological databanks and integrated data analysis tools. In Higgins,D. and Taylor,W. (eds), Bioinformatics—A Practical Approach. Oxford University Press, Oxford, UK, pp. 215–241.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

