The length of telomeric G-rich strand 3'-overhang measured by oligonucleotide ligation assay
- PMID: 11266570
- PMCID: PMC31296
- DOI: 10.1093/nar/29.7.e35
The length of telomeric G-rich strand 3'-overhang measured by oligonucleotide ligation assay
Abstract
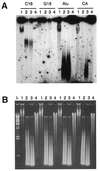
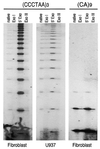
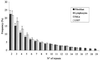
A typical G-rich telomeric DNA strand, which runs 5'-->3' toward the chromosome ends, protrudes by several nucleotides in lower eukaryotes. In human chromosomes long G-rich 3'-overhangs have been found. Apart from the standard G-rich tail, several non-canonical terminal structures have been proposed. However, the mechanism of long-tail formation, the presence and the role of these structures in telomere maintenance or shortening are not completely understood. In a search for a simple method to accurately measure the 3'-overhang we have established a protocol based on the ligation of telomeric oligonucleotide hybridized to non-denatured DNA under stringent conditions (oligonucleotide ligation assay with telomeric repeat oligonucleotide). This method enabled us to detect a large proportion of G-rich single-stranded telomeric DNA that was as short as 24 nt. Nevertheless, we showed G-tails longer than 400 nt. In all tested cells the lengths ranging from 108 to 270 nt represented only 37% of the whole molecule population, while 56-62% were <90 nt. Our protocol provides a simple and sensitive method for measuring the length of naturally occurring unpaired repeated DNA.
Figures

References
-
- Muller H.J. (1938) The remaking of chromosomes. The Collecting Net, Woods Hope (USA), 13, 181–195.
-
- Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. - PubMed
-
- Blackburn E.H. (1994) Telomeres: no end in sight. Cell, 77, 621–623. - PubMed
-
- Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

