Detection of mitochondrial single nucleotide polymorphisms using a primer elongation reaction on oligonucleotide microarrays
- PMID: 11266571
- PMCID: PMC31297
- DOI: 10.1093/nar/29.7.e36
Detection of mitochondrial single nucleotide polymorphisms using a primer elongation reaction on oligonucleotide microarrays
Abstract
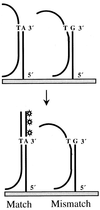
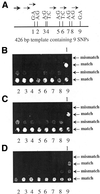
We have developed a novel allele-specific primer elongation protocol using a DNA polymerase on oligonucleotide chips. Oligonucleotide primers carrying polymorphic sites at their free 3'end were covalently bound to glass slides. The generation of single-stranded targets of genomic DNA containing single nuclotide polymorphisms (SNPs) to be typed was achieved by an asymmetric PCR reaction or exonuclease treatment of phosphothioate (PTO)-modified PCR products. In the presence of DNA polymerase and all four dNTPs, with Cy3-dUTP replacing dTTP, allele-specific extension of the immobilized primers took place along a stretch of target DNA sequence. The yield of elongated products was increased by repeated reaction cycles. We performed multiplexed assays with many small DNA targets, or used single targets of up to 4.4 kb mitochondrial DNA (mtDNA) sequence to detect multiple SNPs in one reaction. The latter approach greatly simplifies preamplification of SNP-containing regions, thereby providing a framework for typing hundreds of mtDNA polymorphisms.
Figures




Similar articles
-
SNP typing by apyrase-mediated allele-specific primer extension on DNA microarrays.Nucleic Acids Res. 2002 Aug 1;30(15):e75. doi: 10.1093/nar/gnf074. Nucleic Acids Res. 2002. PMID: 12140337 Free PMC article.
-
[Application of oligonucleotide microarray primer extension to detection of p53 single nucleotide polymorphisms].Ai Zheng. 2005 Jul;24(7):898-902. Ai Zheng. 2005. PMID: 16004824 Chinese.
-
A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays.Genome Res. 2000 Jul;10(7):1031-42. doi: 10.1101/gr.10.7.1031. Genome Res. 2000. PMID: 10899152 Free PMC article.
-
Primer design for multiplexed genotyping.Methods Mol Biol. 2007;402:269-86. doi: 10.1007/978-1-59745-528-2_13. Methods Mol Biol. 2007. PMID: 17951800 Review.
-
Advances in the analysis of DNA sequence variations using oligonucleotide microchip technology.Curr Opin Biotechnol. 2001 Feb;12(1):53-8. doi: 10.1016/s0958-1669(00)00168-3. Curr Opin Biotechnol. 2001. PMID: 11167073 Review.
Cited by
-
SNP typing by apyrase-mediated allele-specific primer extension on DNA microarrays.Nucleic Acids Res. 2002 Aug 1;30(15):e75. doi: 10.1093/nar/gnf074. Nucleic Acids Res. 2002. PMID: 12140337 Free PMC article.
-
TGFA and IRF6 contribute to the risk of nonsyndromic cleft lip with or without cleft palate in northeast China.PLoS One. 2013 Aug 6;8(8):e70754. doi: 10.1371/journal.pone.0070754. Print 2013. PLoS One. 2013. PMID: 23940636 Free PMC article.
-
DNA microarrays with PAMAM dendritic linker systems.Nucleic Acids Res. 2002 Jan 15;30(2):E10. doi: 10.1093/nar/30.2.e10. Nucleic Acids Res. 2002. PMID: 11788736 Free PMC article.
-
An Optimized Preparation Method for Long ssDNA Donors to Facilitate Quick Knock-In Mouse Generation.Cells. 2021 Apr 30;10(5):1076. doi: 10.3390/cells10051076. Cells. 2021. PMID: 33946570 Free PMC article.
-
Visual detection of single-base mismatches in DNA using hairpin oligonucleotide with double-target DNA binding sequences and gold nanoparticles.Biosens Bioelectron. 2012 Apr 15;34(1):37-43. doi: 10.1016/j.bios.2011.12.055. Epub 2012 Feb 10. Biosens Bioelectron. 2012. PMID: 22386491 Free PMC article.
References
-
- Brookes A.J. (1999) The essence of SNPs. Gene, 234, 177–186. - PubMed
-
- Cargill M., Altshuler,D., Ireland,J., Sklar,P., Ardlie,K., Patil,N., Lane,C.R., Lim,E.P., Kalyanaraman,N., Nemesh,J. et al. (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet., 22, 231–238. - PubMed
-
- Halushka M.K., Fan,J.B., Bentley,K., Hsie,L., Shen,N., Weder,A., Cooper,R., Lipshutz,R. and Chakravarti,A. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet., 22, 239–247. - PubMed
-
- Kruglyak L. (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet., 22, 139–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

