Plasticity of quaternary structure: twenty-two ways to form a LacI dimer
- PMID: 11266612
- PMCID: PMC2373939
- DOI: 10.1110/ps.35801
Plasticity of quaternary structure: twenty-two ways to form a LacI dimer
Abstract
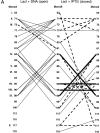
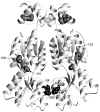
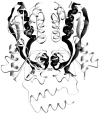
The repressor proteins of the LacI/GalR family exhibit significant similarity in their secondary and tertiary structures despite less than 35% identity in their primary sequences. Furthermore, the core domains of these oligomeric repressors, which mediate dimerization, are homologous with the monomeric periplasmic binding proteins, extending the issue of plasticity to quaternary structure. To elucidate the determinants of assembly, a structure-based alignment has been created for three repressors and four periplasmic binding proteins. Contact maps have also been constructed for the three repressor interfaces to distinguish any conserved interactions. These analyses show few strict requirements for assembly of the core N-subdomain interface. The interfaces of repressor core C-subdomains are well conserved at the structural level, and their primary sequences differ significantly from the monomeric periplasmic binding proteins at positions equivalent to LacI 281 and 282. However, previous biochemical and phenotypic analyses indicate that LacI tolerates many mutations at 281. Mutations at LacI 282 were shown to abrogate assembly, but for Y282D this could be compensated by a second-site mutation in the core N-subdomain at K84 to L or A. Using the link between LacI assembly and function, we have further identified 22 second-site mutations that compensate the Y282D dimerization defect in vivo. The sites of these mutations fall into several structural regions, each of which may influence assembly by a different mechanism. Thus, the 360-amino acid scaffold of LacI allows plasticity of its quaternary structure. The periplasmic binding proteins may require only minimal changes to facilitate oligomerization similar to the repressor proteins.
Figures












Similar articles
-
Fine-tuning function: correlation of hinge domain interactions with functional distinctions between LacI and PurR.Protein Sci. 2002 Apr;11(4):778-94. doi: 10.1110/ps.4050102. Protein Sci. 2002. PMID: 11910022 Free PMC article.
-
Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as "spacers" which do not require a specific sequence.J Mol Biol. 1994 Jul 29;240(5):421-33. doi: 10.1006/jmbi.1994.1458. J Mol Biol. 1994. PMID: 8046748
-
Parallel evolution of ligand specificity between LacI/GalR family repressors and periplasmic sugar-binding proteins.Mol Biol Evol. 2003 Feb;20(2):267-77. doi: 10.1093/molbev/msg038. Mol Biol Evol. 2003. PMID: 12598694
-
Lac repressor genetic map in real space.Trends Biochem Sci. 1997 Sep;22(9):334-9. doi: 10.1016/s0968-0004(97)01104-3. Trends Biochem Sci. 1997. PMID: 9301333 Review.
-
Lac repressor at last.Structure. 1996 Mar 15;4(3):219-22. doi: 10.1016/s0969-2126(96)00025-1. Structure. 1996. PMID: 8805532 Review.
Cited by
-
Data on publications, structural analyses, and queries used to build and utilize the AlloRep database.Data Brief. 2016 Jul 9;8:948-57. doi: 10.1016/j.dib.2016.07.006. eCollection 2016 Sep. Data Brief. 2016. PMID: 27508249 Free PMC article.
-
Fine-tuning function: correlation of hinge domain interactions with functional distinctions between LacI and PurR.Protein Sci. 2002 Apr;11(4):778-94. doi: 10.1110/ps.4050102. Protein Sci. 2002. PMID: 11910022 Free PMC article.
-
Functional consequences of exchanging domains between LacI and PurR are mediated by the intervening linker sequence.Proteins. 2007 Jul 1;68(1):375-88. doi: 10.1002/prot.21412. Proteins. 2007. PMID: 17436321 Free PMC article.
-
A novel molecular switch.J Mol Biol. 2009 Aug 28;391(4):661-70. doi: 10.1016/j.jmb.2009.06.039. Epub 2009 Jun 21. J Mol Biol. 2009. PMID: 19540845 Free PMC article.
-
Engineering an allosteric transcription factor to respond to new ligands.Nat Methods. 2016 Feb;13(2):177-83. doi: 10.1038/nmeth.3696. Epub 2015 Dec 21. Nat Methods. 2016. PMID: 26689263 Free PMC article.
References
-
- Alberti, S., Oehler, S., von Wilcken-Bergmann, B., Krämer, H., and Müller-Hill, B. 1991. Dimer-to-tetramer assembly of Lac repressor involves a leucine heptad repeat. The New Biologist 3 57–62. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Barry, J.K. and Matthews, K.S. 1999. Substitutions at histidine 74 and aspartate 278 alter ligand binding and allostery in lactose repressor protein. Biochemistry 38 3579–3590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

