Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5' nuclease quantitative reverse transcription-polymerase chain reaction
- PMID: 11272893
- PMCID: PMC1906896
- DOI: 10.1016/S1525-1578(10)60621-6
Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5' nuclease quantitative reverse transcription-polymerase chain reaction
Abstract
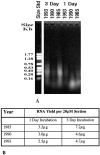
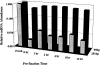
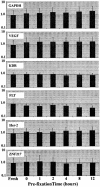
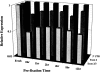
Analysis of gene expression and correlation with clinical parameters has the potential to become an important factor in therapeutic decision making. The ability to analyze gene expression in archived tissues, for which clinical followup is already available, will greatly facilitate research in this area. A major obstacle to this approach, however, has been the uncertainty about whether gene expression analyses from routinely archived tissues accurately reflect expression before fixation. In the present study we have optimized the RNA isolation and reverse transcription steps for quantitative reverse transcription-polymerase chain reaction (RT-PCR) on archival material. Using tissue taken directly from the operating room, mRNAs with half-lives from 10 minutes to >8 hours were isolated and reverse transcribed. Subsequent real-time quantitative PCR methodology (TaqMan) on these cDNAs gives a measurement of gene expression in the fixed tissues comparable to that in the fresh tissue. In addition, we simulated routine pathology handling and demonstrate that this method of mRNA quantitation is insensitive to pre-fixation times (time from excision to fixation) of up to 12 hours. Therefore, it should be feasible to analyze gene expression in archived tissues where tissue collection procedures are largely unknown.
Figures

References
-
- Fearon ER, Dang CV: Cancer genetics: tumor suppressor meets oncogene. Curr Biol 1999, 9:R62-R65 - PubMed
-
- Hill JR, Kuriyama N, Kuriyama H, Israel MA: Molecular genetics of brain tumors. Arch Neurol 1999, 56:439-441 - PubMed
-
- Pearson PL, Van der Luijt RB: The genetic analysis of cancer. J Intern Med 1998, 243:413-417 - PubMed
-
- Steele A, Uckan D, Steele P, Chamizo W, Washington K, Koutsonikolis A, Good RA: RT in situ PCR for the detection of mRNA transcripts of Fas-L in the immune-privileged placental environment. Cell Vis 1998, 5:13-19 - PubMed
-
- Gruber AD, Greiser-Wilke IM, Haas L, Hewicker-Trautwein M, Moennig V: Detection of bovine viral diarrhea virus RNA in formalin-fixed, paraffin-embedded brain tissue by nested polymerase chain reaction. J Virol Methods 1993, 43:309-319 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

