Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development
- PMID: 11274056
- PMCID: PMC312657
- DOI: 10.1101/gad.194101
Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development
Abstract
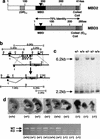
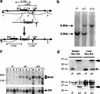
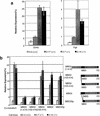
MBD2 and MBD3 are closely related proteins with consensus methyl-CpG binding domains. MBD2 is a transcriptional repressor that specifically binds to methylated DNA and is a component of the MeCP1 protein complex. In contrast, MBD3 fails to bind methylated DNA in murine cells, and is a component of the Mi-2/NuRD corepressor complex. We show by gene targeting that the two proteins are not functionally redundant in mice, as Mbd3-/- mice die during early embryogenesis, whereas Mbd2-/- mice are viable and fertile. Maternal behavior of Mbd2-/- mice is however defective and, at the molecular level, Mbd2-/- mice lack a component of MeCP1. Mbd2-mutant cells fail to fully silence transcription from exogenous methylated templates, but inappropriate activation of endogenous imprinted genes or retroviral sequences was not detected. Despite their differences, Mbd3 and Mbd2 interact genetically suggesting a functional relationship. Genetic and biochemical data together favor the view that MBD3 is a key component of the Mi-2/NuRD corepressor complex, whereas MBD2 may be one of several factors that can recruit this complex to DNA.
Figures


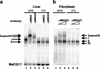

References
-
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. - PubMed
-
- Amir RE, Van Den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. - PubMed
-
- Antequera F, Macleod D, Bird AP. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. - PubMed
-
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. - PubMed
-
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
