Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different
- PMID: 11274097
- PMCID: PMC95154
- DOI: 10.1128/JB.183.8.2405-2410.2001
Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different
Abstract
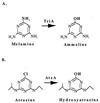
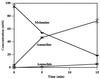
The gene encoding melamine deaminase (TriA) from Pseudomonas sp. strain NRRL B-12227 was identified, cloned into Escherichia coli, sequenced, and expressed for in vitro study of enzyme activity. Melamine deaminase displaced two of the three amino groups from melamine, producing ammeline and ammelide as sequential products. The first deamination reaction occurred more than 10 times faster than the second. Ammelide did not inhibit the first or second deamination reaction, suggesting that the lower rate of ammeline hydrolysis was due to differential substrate turnover rather than product inhibition. Remarkably, melamine deaminase is 98% identical to the enzyme atrazine chlorohydrolase (AtzA) from Pseudomonas sp. strain ADP. Each enzyme consists of 475 amino acids and differs by only 9 amino acids. AtzA was shown to exclusively catalyze dehalogenation of halo-substituted triazine ring compounds and had no activity with melamine and ammeline. Similarly, melamine deaminase had no detectable activity with the halo-triazine substrates. Melamine deaminase was active in deamination of a substrate that was structurally identical to atrazine, except for the substitution of an amino group for the chlorine atom. Moreover, melamine deaminase and AtzA are found in bacteria that grow on melamine and atrazine compounds, respectively. These data strongly suggest that the 9 amino acid differences between melamine deaminase and AtzA represent a short evolutionary pathway connecting enzymes catalyzing physiologically relevant deamination and dehalogenation reactions, respectively.
Figures


References
-
- Akashi H. Within- and between-species DNA sequence variation and the ‘footprint’ of natural selection. Gene. 1999;238:39–51. - PubMed
-
- Alvey S, Crowley D E. Influence of organic amendments on biodegradation of atrazine as a nitrogen source. J Environ Qual. 1995;24:1156–1162.
-
- Belluck D A, Benjamin S L, Dawson T. Groundwater contamination by atrazine and its metabolites: risk assessment, policy and legal implications. In: Somasundaram L, Coats J R, editors. Pesticide transformation products: fate and significance in the environment. Washington, D.C.: American Chemical Society; 1991. pp. 254–273.
-
- Benning M M, Kuo J M, Raushel F M, Holden H M. Three-dimensional structure of phosphotriesterase: an enzyme capable of detoxifying organophosphate nerve agents. Biochemistry. 1994;33:15001–15007. - PubMed
-
- Benning M M, Kuo J M, Raushel F M, Holden H M. Three-dimensional structure of the binuclear metal center of phosphotriesterase. Biochemistry. 1995;34:7973–7978. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

