Independence of circadian timing from cell division in cyanobacteria
- PMID: 11274102
- PMCID: PMC95159
- DOI: 10.1128/JB.183.8.2439-2444.2001
Independence of circadian timing from cell division in cyanobacteria
Abstract
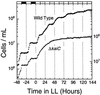
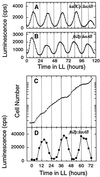
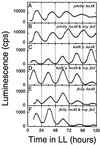
In the cyanobacterium Synechococcus elongatus, cell division is regulated by a circadian clock. Deletion of the circadian clock gene, kaiC, abolishes rhythms of gene expression and cell division timing. Overexpression of the ftsZ gene halted cell division but not growth, causing cells to grow as filaments without dividing. The nondividing filamentous cells still exhibited robust circadian rhythms of gene expression. This result indicates that the circadian timing system is independent of rhythmic cell division and, together with other results, suggests that the cyanobacterial circadian system is stable and well sustained under a wide range of intracellular conditions.
Figures


References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andersson C R, Tsinoremas N F, Shelton J, Lebedeva N V, Yarrow J, Min H, Golden S S. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. - PubMed
-
- Barkai N, Leibler S. Biological rhythms: circadian clocks limited by noise. Nature. 2000;403:267–268. - PubMed
-
- Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
-
- Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

