Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis
- PMID: 11274109
- PMCID: PMC95166
- DOI: 10.1128/JB.183.8.2497-2504.2001
Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis
Abstract
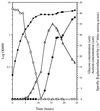
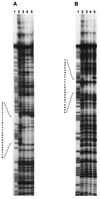
Bacillus subtilis grown in media containing amino acids or glucose secretes acetate, pyruvate, and large quantities of acetoin into the growth medium. Acetoin can be reused by the bacteria during stationary phase when other carbon sources have been depleted. The acoABCL operon encodes the E1alpha, E1beta, E2, and E3 subunits of the acetoin dehydrogenase complex in B. subtilis. Expression of this operon is induced by acetoin and repressed by glucose in the growth medium. The acoR gene is located downstream from the acoABCL operon and encodes a positive regulator which stimulates the transcription of the operon. The product of acoR has similarities to transcriptional activators of sigma 54-dependent promoters. The four genes of the operon are transcribed from a -12, -24 promoter, and transcription is abolished in acoR and sigL mutants. Deletion analysis showed that DNA sequences more than 85 bp upstream from the transcriptional start site are necessary for full induction of the operon. These upstream activating sequences are probably the targets of AcoR. Analysis of an acoR'-'lacZ strain of B. subtilis showed that the expression of acoR is not induced by acetoin and is repressed by the presence of glucose in the growth medium. Transcription of acoR is also negatively controlled by CcpA, a global regulator of carbon catabolite repression. A specific interaction of CcpA in the upstream region of acoR was demonstrated by DNase I footprinting experiments, suggesting that repression of transcription of acoR is mediated by the binding of CcpA to the promoter region of acoR.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

