Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri
- PMID: 11274111
- PMCID: PMC95168
- DOI: 10.1128/JB.183.8.2516-2526.2001
Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri
Abstract
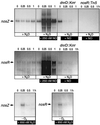
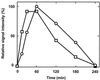
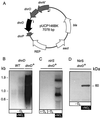
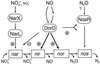
The expression of denitrification by a facultatively anaerobic bacterium requires as exogenous signals a low oxygen tension concomitant with an N oxide. We have studied the role of nitric oxide (NO), nitrous oxide (N2O), and nitrite as signal molecules for the expression of the denitrification apparatus of Pseudomonas stutzeri. Transcriptional kinetics of structural genes were monitored by Northern blot analysis in a 60-min time frame after cells were exposed to an N oxide signal. To differentiate the inducer role of NO from that of nitrite, mRNA kinetics were monitored under anoxic conditions in a nirF strain, where NO generation from nitrite is prevented because of a defect in heme D(1) biosynthesis. NO-triggered responses were monitored from the nirSTB operon (encoding cytochrome cd(1) nitrite reductase), the norCB operon (encoding NO reductase), nosZ (encoding nitrous oxide reductase), and nosR (encoding a putative regulator). Transcription of nirSTB and norCB was activated by 5 to 50 nM NO, whereas the nosZ promoter required about 250 nM. Nitrite at 5 to 50 nM elicited no response. At a threshold concentration of 650 nM N2O, we observed in the anoxic cell the transient appearance of nosZ and nosR transcripts. Constant levels of transcripts of both genes were observed in an anoxic cell sparged with N2O. NO at 250 nM stimulated in this cell type the expression of nos genes severalfold. The transcription factor DnrD, a member of the FNR-CRP family, was found to be part of the NO-triggered signal transduction pathway. However, overexpression of dnrD in an engineered strain did not result in NirS synthesis, indicating a need for activation of DnrD. NO modified the transcriptional pattern of the dnrD operon by inducing the transcription of dnrN and dnrO, located upstream of dnrD. Insertional mutagenesis of dnrN altered the kinetic response of the nirSTB operon towards nitrite. Our data establish NO and DnrD as key elements in the regulatory network of denitrification in P. stutzeri. The NO response adds to the previously identified nitrate-nitrite response mediated by the NarXL two-component system for the expression of respiratory nitrate reductase encoded by the narGHJI operon.
Figures




References
-
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:11905–11910. - PubMed
-
- Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–55. - PubMed
-
- Arai H, Igarashi Y, Kodama T. The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1995;1261:279–284. - PubMed
-
- Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. - PubMed
-
- Arai H, Kodama T, Igarashi Y. Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;170:19–24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

