Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus
- PMID: 11274122
- PMCID: PMC95179
- DOI: 10.1128/JB.183.8.2614-2623.2001
Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus
Abstract
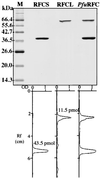
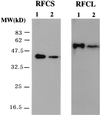
Replication factor C (RFC) and proliferating cell nuclear antigen (PCNA) are accessory proteins essential for processive DNA synthesis in the domain Eucarya. The function of RFC is to load PCNA, a processivity factor of eukaryotic DNA polymerases delta and epsilon, onto primed DNA templates. RFC-like genes, arranged in tandem in the Pyrococcus furiosus genome, were cloned and expressed individually in Escherichia coli cells to determine their roles in DNA synthesis. The P. furiosus RFC (PfuRFC) consists of a small subunit (RFCS) and a large subunit (RFCL). Highly purified RFCS possesses an ATPase activity, which was stimulated up to twofold in the presence of both single-stranded DNA (ssDNA) and P. furiosus PCNA (PfuPCNA). The ATPase activity of PfuRFC itself was as strong as that of RFCS. However, in the presence of PfuPCNA and ssDNA, PfuRFC exhibited a 10-fold increase in ATPase activity under the same conditions. RFCL formed very large complexes by itself and had an extremely weak ATPase activity, which was not stimulated by PfuPCNA and DNA. The PfuRFC stimulated PfuPCNA-dependent DNA synthesis by both polymerase I and polymerase II from P. furiosus. We propose that PfuRFC is required for efficient loading of PfuPCNA and that the role of RFC in processive DNA synthesis is conserved in Archaea and Eucarya.
Figures




References
-
- Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O'Donnell M, Hurwitz J. A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem. 1997;272:18974–18981. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

