Protein folding from a highly disordered denatured state: the folding pathway of chymotrypsin inhibitor 2 at atomic resolution
- PMID: 11274353
- PMCID: PMC31838
- DOI: 10.1073/pnas.071054398
Protein folding from a highly disordered denatured state: the folding pathway of chymotrypsin inhibitor 2 at atomic resolution
Abstract
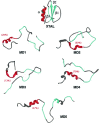
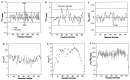
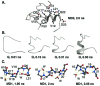
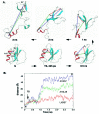
Previous experimental and theoretical studies have produced high-resolution descriptions of the native and folding transition states of chymotrypsin inhibitor 2 (CI2). In similar fashion, here we use a combination of NMR experiments and molecular dynamics simulations to examine the conformations populated by CI2 in the denatured state. The denatured state is highly unfolded, but there is some residual native helical structure along with hydrophobic clustering in the center of the chain. The lack of persistent nonnative structure in the denatured state reduces barriers that must be overcome, leading to fast folding through a nucleation-condensation mechanism. With the characterization of the denatured state, we have now completed our description of the folding/unfolding pathway of CI2 at atomic resolution.
Figures

References
-
- Wong K B, Clarke J, Bond C J, Neira J L, Freund S M V, Fersht A R F, Daggett V. J Mol Biol. 2000;296:1257–1282. - PubMed
-
- Matouschek A, Kellis J T, Jr, Serrano L, Fersht A R. Nature (London) 1989;340:122–126. - PubMed
-
- Li A, Daggett V. J Mol Biol. 1996;257:412–429. - PubMed
-
- Daggett V, Li A, Itzhaki L S, Otzen D E, Fersht A R. J Mol Biol. 1996;257:430–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

