Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells
- PMID: 11274400
- PMCID: PMC31137
- DOI: 10.1073/pnas.071043198
Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells
Abstract
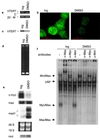
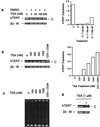
Recent evidence suggests that the Myc and Mad1 proteins are implicated in the regulation of the gene encoding the human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase. We have analyzed the in vivo interaction between endogenous c-Myc and Mad1 proteins and the hTERT promoter in HL60 cells with the use of the chromatin immunoprecipitation assay. The E-boxes at the hTERT proximal promoter were occupied in vivo by c-Myc in exponentially proliferating HL60 cells but not in cells induced to differentiate by DMSO. In contrast, Mad1 protein was induced and bound to the hTERT promoter in differentiated HL60 cells. Concomitantly, the acetylation of the histones at the promoter was significantly reduced. These data suggest that the reciprocal E-box occupancy by c-Myc and Mad1 is responsible for activation and repression of the hTERT gene in proliferating and differentiated HL60 cells, respectively. Furthermore, the histone deacetylase inhibitor trichostatin A inhibited deacetylation of histones at the hTERT promoter and attenuated the repression of hTERT transcription during HL60 cell differentiation. In addition, trichostatin A treatment activated hTERT transcription in resting human lymphocytes and fibroblasts. Taken together, these results indicate that acetylation/deacetylation of histones is operative in the regulation of hTERT expression.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

