Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha
- PMID: 11274403
- PMCID: PMC31140
- DOI: 10.1073/pnas.061036798
Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha
Abstract
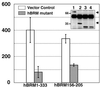
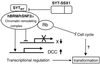
Human synovial sarcoma has been shown to exclusively harbor the chromosomal translocation t(X;18) that produces the chimeric gene SYT-SSX. However, the role of SYT-SSX in cellular transformation remains unclear. In this study, we have established 3Y1 rat fibroblast cell lines that constitutively express SYT, SSX1, and SYT-SSX1 and found that SYT-SSX1 promoted growth rate in culture, anchorage-independent growth in soft agar, and tumor formation in nude mice. Deletion of the N-terminal 181 amino acids of SYT-SSX1 caused loss of its transforming activity. Furthermore, association of SYT-SSX1 with the chromatin remodeling factor hBRM/hSNF2 alpha, which regulates transcription, was demonstrated in both SYT-SSX1-expressing 3Y1 cells and in the human synovial sarcoma cell line HS-SY-II. The binding region between the two molecules was shown to reside within the N-terminal 181 amino acids stretch (aa 1--181) of SYT-SSX1 and 50 amino acids (aa 156--205) of hBRM/hSNF2 alpha and we found that the overexpression of this binding region of hBRM/hSNF2 alpha significantly suppressed the anchorage-independent growth of SYT-SSX1-expressing 3Y1 cells. To analyze the transcriptional regulation by SYT-SSX1, we established conditional expression system of SYT-SSX1 and examined the gene expression profiles. The down-regulation of potential tumor suppressor DCC was observed among 1,176 genes analyzed by microarray analysis, and semi-quantitative reverse transcription--PCR confirmed this finding. These data clearly demonstrate transforming activity of human oncogene SYT-SSX1 and also involvement of chromatin remodeling factor hBRM/hSNF2 alpha in human cancer.
Figures


References
-
- Enzinger F M, Weiss S W. Soft Tissue Tumors. St. Louis: Mosby; 1988.
-
- Wang-Wuu S, Soukop S W, Lange B J. Cancer Genet Cytogenet. 1987;29:179–181. - PubMed
-
- Nojima T, Wang Y S, Abe S, Matsuno T, Yamawaki S, Nagashima K. Acta Pathol Jpn. 1990;40:486–493. - PubMed
-
- Knight J C, Reeves B R, Kearney L, Monaco A P, Lehrach H, Cooper C S. Int J Oncol. 1992;1:747–752. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

