Microbial phyllosphere populations are more complex than previously realized
- PMID: 11274410
- PMCID: PMC31148
- DOI: 10.1073/pnas.051633898
Microbial phyllosphere populations are more complex than previously realized
Abstract
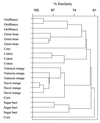
Phyllosphere microbial communities were evaluated on leaves of field-grown plant species by culture-dependent and -independent methods. Denaturing gradient gel electrophoresis (DGGE) with 16S rDNA primers generally indicated that microbial community structures were similar on different individuals of the same plant species, but unique on different plant species. Phyllosphere bacteria were identified from Citrus sinesis (cv. Valencia) by using DGGE analysis followed by cloning and sequencing of the dominant rDNA bands. Of the 17 unique sequences obtained, database queries showed only four strains that had been described previously as phyllosphere bacteria. Five of the 17 sequences had 16S similarities lower than 90% to database entries, suggesting that they represent previously undescribed species. In addition, three fungal species were also identified. Very different 16S rDNA DGGE banding profiles were obtained when replicate cv. Valencia leaf samples were cultured in BIOLOG EcoPlates for 4.5 days. All of these rDNA sequences had 97--100% similarity to those of known phyllosphere bacteria, but only two of them matched those identified by the culture independent DGGE analysis. Like other studied ecosystems, microbial phyllosphere communities therefore are more complex than previously thought, based on conventional culture-based methods.
Figures


References
-
- Beattie G A, Lindow S E. Phytopathology. 1999;89:353–359. - PubMed
-
- Pattantyus J, Kiss S. Stud Univ Babes-Bolyai Biol. 1994;39:97–103.
-
- Hallmann J, Quadt-Hallmann A, Mahaffee W F, Kloepper J W. Can J Microbiol. 1997;43:895–914.
-
- Andrews J H, Kenerley C M, Nordheim E V. Microb Ecol. 1980;6:71–84. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

