Strain-specified relative conformational stability of the scrapie prion protein
- PMID: 11274476
- PMCID: PMC2373967
- DOI: 10.1110/ps.39201
Strain-specified relative conformational stability of the scrapie prion protein
Abstract
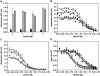
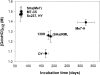
Studies of prion biology and diseases have elucidated several new concepts, but none was more heretical than the proposal that the biological properties that distinguish different prion strains are enciphered in the disease-causing prion protein (PrP(Sc)). To explore this postulate, we examined the properties of PrP(Sc) from eight prion isolates that propagate in Syrian hamster (SHa). Using resistance to protease digestion as a marker for the undenatured protein, we examined the conformational stabilities of these PrP(Sc) molecules. All eight isolates showed sigmoidal patterns of transition from native to denatured PrP(Sc) as a function of increasing guanidine hydrochloride (GdnHCl) concentration. Half-maximal denaturation occurred at a mean value of 1.48 M GdnHCl for the Sc237, HY, SHa(Me7), and MT-C5 isolates, all of which have approximately 75-d incubation periods; a concentration of 1.08 M was found for the DY strain with a approximately 170-d incubation period and approximately 1.25 M for the SHa(RML) and 139H isolates with approximately 180-d incubation periods. A mean value of 1.39 M GdnHCl for the Me7-H strain with a approximately 320-d incubation period was found. Based on these results, the eight prion strains segregated into four distinct groups. Our results support the unorthodox proposal that distinct PrP(Sc) conformers encipher the biological properties of prion strains.
Figures
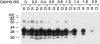

References
-
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181 223–230. - PubMed
-
- Bellinger-Kawahara, C.G., Kempner, E., Groth, D.F., Gabizon, R., and Prusiner, S.B. 1988. Scrapie prion liposomes and rods exhibit target sizes of 55,000 Da. Virology 164 537–541. - PubMed
-
- ———. 1992b. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 73 329–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

