The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins
- PMID: 11278557
- PMCID: PMC7982318
- DOI: 10.1074/jbc.M009731200
The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins
Abstract
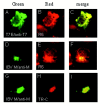
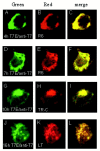
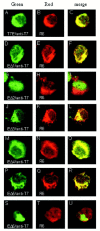
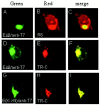
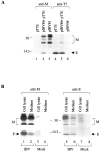
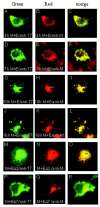
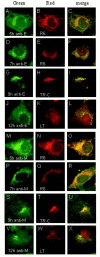
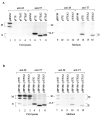
One missing link in the coronavirus assembly is the physical interaction between two crucial structural proteins, the membrane (M) and envelope (E) proteins. In this study, we demonstrate that the coronavirus infectious bronchitis virus E can physically interact, via a putative peripheral domain, with M. Deletion of this domain resulted in a drastic reduction in the incorporation of M into virus-like particles. Immunofluorescent staining of cells coexpressing M and E supports that E interacts with M and relocates M to the same subcellular compartments that E resides in. E was retained in the pre-Golgi membranes, prior to being translocated to the Golgi apparatus and the secretory vesicles; M was observed to exhibit similar localization and translocation profiles as E when coexpressed with E. Deletion studies identified the C-terminal 6-residue RDKLYS as the endoplasmic reticulum retention signal of E, and site-directed mutagenesis of the -4 lysine residue to glutamine resulted in the accumulation of E in the Golgi apparatus. The third domain of E that plays a crucial role in virus budding is a putative transmembrane domain present at the N-terminal region, because deletion of the domain resulted in a free distribution of the mutant protein and in dysfunctional viral assembly.
Figures


Similar articles
-
Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles.J Virol. 2000 May;74(9):4319-26. doi: 10.1128/jvi.74.9.4319-4326.2000. J Virol. 2000. PMID: 10756047 Free PMC article.
-
The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles.J Biol Chem. 2021 Jan-Jun;296:100111. doi: 10.1074/jbc.RA120.016175. Epub 2020 Dec 3. J Biol Chem. 2021. PMID: 33229438 Free PMC article.
-
The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus.J Virol. 2019 May 15;93(11):e00015-19. doi: 10.1128/JVI.00015-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30867314 Free PMC article.
-
Self-assembly of severe acute respiratory syndrome coronavirus membrane protein.J Biol Chem. 2010 Apr 23;285(17):12862-72. doi: 10.1074/jbc.M109.030270. Epub 2010 Feb 12. J Biol Chem. 2010. PMID: 20154085 Free PMC article.
-
Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein.Virol J. 2009 Jun 18;6:79. doi: 10.1186/1743-422X-6-79. Virol J. 2009. PMID: 19534833 Free PMC article.
Cited by
-
The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles.J Gen Virol. 2016 Aug;97(8):1853-1864. doi: 10.1099/jgv.0.000494. Epub 2016 May 4. J Gen Virol. 2016. PMID: 27145752 Free PMC article.
-
Incorporation of spike and membrane glycoproteins into coronavirus virions.Viruses. 2015 Apr 3;7(4):1700-25. doi: 10.3390/v7041700. Viruses. 2015. PMID: 25855243 Free PMC article. Review.
-
Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity.Proc Natl Acad Sci U S A. 2004 Jun 29;101(26):9804-9. doi: 10.1073/pnas.0403492101. Epub 2004 Jun 21. Proc Natl Acad Sci U S A. 2004. PMID: 15210961 Free PMC article.
-
Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells.FEBS Lett. 2006 May 29;580(13):3192-200. doi: 10.1016/j.febslet.2006.04.076. Epub 2006 May 4. FEBS Lett. 2006. PMID: 16684538 Free PMC article.
-
Coronavirus envelope protein: current knowledge.Virol J. 2019 May 27;16(1):69. doi: 10.1186/s12985-019-1182-0. Virol J. 2019. PMID: 31133031 Free PMC article. Review.
References
-
- Brautigam S., Snezhkov E., Bishop D.H. Virology. 1993;192:512–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

