Determination of DNA content of aquatic bacteria by flow cytometry
- PMID: 11282616
- PMCID: PMC92780
- DOI: 10.1128/AEM.67.4.1636-1645.2001
Determination of DNA content of aquatic bacteria by flow cytometry
Abstract
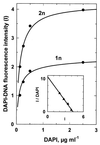
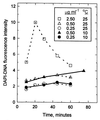
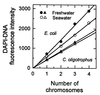
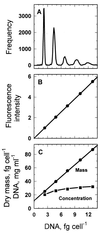
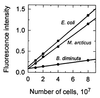
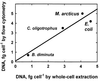
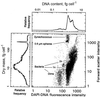
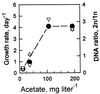
The distribution of DNA among bacterioplankton and bacterial isolates was determined by flow cytometry of DAPI (4',6'-diamidino-2-phenylindole)-stained organisms. Conditions were optimized to minimize error from nonspecific staining, AT bias, DNA packing, changes in ionic strength, and differences in cell permeability. The sensitivity was sufficient to characterize the small 1- to 2-Mb-genome organisms in freshwater and seawater, as well as low-DNA cells ("dims"). The dims could be formed from laboratory cultivars; their apparent DNA content was 0.1 Mb and similar to that of many particles in seawater. Preservation with formaldehyde stabilized samples until analysis. Further permeabilization with Triton X-100 facilitated the penetration of stain into stain-resistant lithotrophs. The amount of DNA per cell determined by flow cytometry agreed with mean values obtained from spectrophotometric analyses of cultures. Correction for the DNA AT bias of the stain was made for bacterial isolates with known G+C contents. The number of chromosome copies per cell was determined with pure cultures, which allowed growth rate analyses based on cell cycle theory. The chromosome ratio was empirically related to the rate of growth, and the rate of growth was related to nutrient concentration through specific affinity theory to obtain a probe for nutrient kinetics. The chromosome size of a Marinobacter arcticus isolate was determined to be 3.0 Mb by this method. In a typical seawater sample the distribution of bacterial DNA revealed two major populations based on DNA content that were not necessarily similar to populations determined by using other stains or protocols. A mean value of 2.5 fg of DNA cell(-1) was obtained for a typical seawater sample, and 90% of the population contained more than 1.1 fg of DNA cell(-1).
Figures


References
-
- Albert F G, Eckdahl T T, Fitzgerald D J, Anderson J N. Heterogeneity in the actions of drugs that bind in the DNA minor groove. Biochemistry. 1999;38:10135–10146. - PubMed
-
- Barcellona M L, Favilla R, VonBerger J, Avitabile M, Ragusa N, Masotti L. DNA–4′-6-diamidino-2-phenylindole interactions: a comparative study employing fluorescence and ultraviolet spectroscopy. Arch Biochem Biophys. 1986;250:48–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

