Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates
- PMID: 11283056
- PMCID: PMC87939
- DOI: 10.1128/JCM.39.4.1353-1359.2001
Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates
Abstract
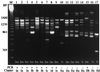
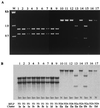
Campylobacter jejuni is one of the most common causes of bacterial diarrhea worldwide and is the primary bacterial cause of food-borne illness. Adherence to and invasion of epithelial cells are the most important pathogenic mechanisms of Campylobacter diarrhea. Molecular characterization of invasive and noninvasive Campylobacter isolates from children with diarrhea and symptom-free children was performed by random amplified polymorphic DNA techniques (RAPD). A distinct RAPD profile with a DNA band of 1.6 kb was observed significantly more frequently among invasive (63%) than among noninvasive (16%) Campylobacter isolates (P = 0.000005). The 1.6-kb band was named the invasion-associated marker (IAM). Using specifically designed primers, a fragment of 518 bp of the iam locus was amplified in 85% of invasive and 20% of noninvasive strains (P = 0.0000000). Molecular typing with a PCR-restriction fragment length polymorphism assay which amplified the entire iam locus showed a HindIII restriction fragment polymorphism pattern associated mainly with invasive strains. Although cluster analysis of the RAPD fingerprinting showed genetic diversity among strains, two main clusters were identified. Cluster I comprised significantly more pathogenic and invasive isolates, while cluster II grouped the majority of nonpathogenic, noninvasive isolates. These data indicate that most of the invasive Campylobacter strains could be differentiated from noninvasive isolates by RAPD analysis and PCR using specific primers that amplify a fragment of the iam locus.
Figures


References
-
- Burucoa C, Frémaux C, Pei Z, Tummuru M, Blaser M J, Cenatiempo Y, Fauchère J L. Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res Microbiol. 1995;146:467–476. - PubMed
-
- Calva J J, Ruiz-Palacios G M, Lopez-Vidal A B, Ramos A, Bojalil R. Cohort study of intestinal infection with Campylobacter in Mexican children. Lancet. 1988;i:503–506. - PubMed
-
- Darling W M, Peel R N, Skirrow M B. Campylobacter cholecystitis. Lancet. 1979;i:1302. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

