Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O
- PMID: 11283059
- PMCID: PMC87942
- DOI: 10.1128/JCM.39.4.1378-1384.2001
Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O
Abstract
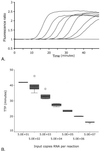
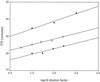
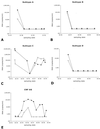
Because human immunodeficiency virus type 1 (HIV-1) subtypes and circulating recombinant forms (CRFs) are spreading rapidly worldwide and are becoming less confined to a geographical area, RNA assays that can detect and quantify all HIV-1 isolates reliably are in demand. We have developed a fast, real-time monitored RNA assay based on an isothermal nucleic acid sequence-based amplification technology that amplifies a part of the long terminal repeat region of the HIV-1 genome. Real-time detection was possible due to the addition of molecular beacons to the amplification reaction that was monitored in a fluorimeter with a thermostat. The lower level of detection of the assay was 10 HIV-1 RNA molecules per reaction, and the lower level of quantification was 100 copies of HIV-1 RNA with a dynamic range of linear quantification between 10(2) and 10(7) RNA molecules. All HIV-1 groups, subtypes, and CRFs could be detected and quantified with equal efficiency, including the group N isolate YBF30 and the group O isolate ANT70. To test the clinical utility of the assay, a series of 62 serum samples containing viruses that encompassed subtypes A through G and CRFs AE and AG of HIV-1 group M were analyzed, and these results were compared to the results of a commercially available assay. This comparison showed that the quantification results correlated highly (R(2) = 0.735) for those subtypes that could be well quantified by both assays (subtypes B, C, D, and F), whereas improved quantification was obtained for subtypes A and G and CRFs AE and AG. A retrospective study with six individuals infected with either a subtype A, B, C, or D or an AG isolate of HIV-1 group M, who were treated with highly active antiretroviral therapy, revealed that the assay was well suited to the monitoring of therapy effects. In conclusion, the newly developed real-time monitored HIV-1 assay is a fast and sensitive assay with a large dynamic range of quantification and is suitable for quantification of most if not all subtypes and groups of HIV-1.
Figures

References
-
- Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. - PubMed
-
- Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J Med Virol. 1996;50:293–302. - PubMed
-
- de Baar M P, De Ronde A, Berkhout B, Cornelissen M, van der Horn K H M, van der Schoot A M, De Wolf F, Lukashov V V, Goudsmit J. Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer-binding site. AIDS Res Hum Retroviruses. 2000;16:499–504. - PubMed
-
- de Baar M P, Janssens W, De Ronde A, Fransen K, Colebunders R, Kestens L, van der Groen G, Goudsmit J. Natural residues versus antiretroviral drug-selected mutations in HIV type 1 group O reverse transcriptase and protease related to virological drug failure in vivo. AIDS Res Hum Retroviruses. 2000;16:1385–1394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

