Structure and specificity of GATA proteins in Th2 development
- PMID: 11283251
- PMCID: PMC86902
- DOI: 10.1128/MCB.21.8.2716-2725.2001
Structure and specificity of GATA proteins in Th2 development
Abstract
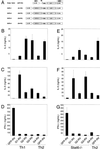
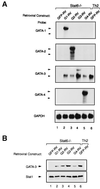
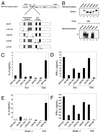
Development of Th2 subset of CD4+ T cells involves the interleukin-4 (IL-4)- and Stat6-dependent increase in GATA-3 expression during primary activation. Recently we reported that the phenotypic stability and factor independence of Th2 cells involves acquisition of an intracellular pathway that maintains GATA-3 expression. Evidence from retroviral expression studies implied that this pathway involved an autoactivation of GATA-3 expression, since Stat6-deficient T cells induced endogenous GATA-3 when infected with GATA-3-expressing retroviruses. That study left unresolved the issue of whether GATA-3 autoactivation was direct or indirect. Several other Th2-specific transcription factors have been described, including c-Maf and JunB. We therefore examined the ability of these other transcription factors to induce GATA-3 expression and promote Th2 development. Neither c-Maf nor JunB induced Th2 development in Stat6-deficient CD4+ T cells, in contrast to GATA-3. Consistent with this indication of a possible direct autoactivation pathway, we also observed that heterologous GATA family proteins GATA-1, GATA-2, and GATA-4 were also capable of inducing GATA-3 expression in developing Stat6-deficient T cells and promote Th2 development. Mutational analysis revealed evidence for two distinct mechanisms of GATA-3 action. IL-4 induction by GATA-3 required each of the functional domains to be present, whereas repression of gamma interferon could occur even when mutants of GATA-3 lacking the second transactivation domain, TA2, were expressed. The GATA-dependent induction of the GATA-3 but not the other GATA genes in T cells suggests that T-cell-specific cis elements within the GATA-3 locus likely cooperate with a general GATA recognition motif to allow GATA-3-dependent autoactivation.
Figures





Similar articles
-
The function role of GATA-3 in Th1 and Th2 differentiation.Immunol Res. 2003;28(1):25-37. doi: 10.1385/IR:28:1:25. Immunol Res. 2003. PMID: 12947222 Review.
-
Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment.Immunity. 2000 Jan;12(1):27-37. doi: 10.1016/s1074-7613(00)80156-9. Immunity. 2000. PMID: 10661403
-
Inhibition of Th2 differentiation and GATA-3 expression by BCL-6.J Immunol. 2003 Mar 1;170(5):2435-41. doi: 10.4049/jimmunol.170.5.2435. J Immunol. 2003. PMID: 12594267
-
Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells.Immunity. 1999 Dec;11(6):677-88. doi: 10.1016/s1074-7613(00)80142-9. Immunity. 1999. PMID: 10626890
-
GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors.Cell Res. 2006 Jan;16(1):3-10. doi: 10.1038/sj.cr.7310002. Cell Res. 2006. PMID: 16467870 Review.
Cited by
-
The function role of GATA-3 in Th1 and Th2 differentiation.Immunol Res. 2003;28(1):25-37. doi: 10.1385/IR:28:1:25. Immunol Res. 2003. PMID: 12947222 Review.
-
The mediatory role of Majie cataplasm on inflammation of allergic asthma through transcription factors related to Th1 and Th2.Chin Med. 2020 May 24;15:53. doi: 10.1186/s13020-020-00334-w. eCollection 2020. Chin Med. 2020. PMID: 32489402 Free PMC article.
-
rVista for comparative sequence-based discovery of functional transcription factor binding sites.Genome Res. 2002 May;12(5):832-9. doi: 10.1101/gr.225502. Genome Res. 2002. PMID: 11997350 Free PMC article.
-
Preliminary Interpretations of Epigenetic Profiling of Cord Blood in Preeclampsia.Genes (Basel). 2022 May 16;13(5):888. doi: 10.3390/genes13050888. Genes (Basel). 2022. PMID: 35627272 Free PMC article.
-
NKG2A and CD56 are coexpressed on activated TH2 but not TH1 lymphocytes.Hum Immunol. 2005 Dec;66(12):1223-34. doi: 10.1016/j.humimm.2006.02.005. Epub 2006 Mar 27. Hum Immunol. 2005. PMID: 16690409 Free PMC article.
References
-
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. - PubMed
-
- Dasen J S, O'Connell S M, Flynn S E, Treier M, Gleiberman A S, Szeto D P, Hooshmand F, Aggarwal A K, Rosenfeld M G. Reciprocal interactions of Pit1 and GATA2 mediate signaling gradient-induced determination of pituitary cell types. Cell. 1999;97:587–598. - PubMed
-
- Dorfman D M, Wilson D B, Bruns G A, Orkin S H. Human transcription factor GATA-2: evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
