Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites
- PMID: 11283258
- PMCID: PMC86909
- DOI: 10.1128/MCB.21.8.2790-2801.2001
Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites
Abstract
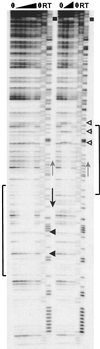
While many of the proteins involved in the initiation of DNA replication are conserved between yeasts and metazoans, the structure of the replication origins themselves has appeared to be different. As typified by ARS1, replication origins in Saccharomyces cerevisiae are <150 bp long and have a simple modular structure, consisting of a single binding site for the origin recognition complex, the replication initiator protein, and one or more accessory sequences. DNA replication initiates from a discrete site. While the important sequences are currently less well defined, metazoan origins appear to be different. These origins are large and appear to be composed of multiple, redundant elements, and replication initiates throughout zones as large as 55 kb. In this report, we characterize two S. cerevisiae replication origins, ARS101 and ARS310, which differ from the paradigm. These origins contain multiple, redundant binding sites for the origin recognition complex. Each binding site must be altered to abolish origin function, while the alteration of a single binding site is sufficient to inactivate ARS1. This redundant structure may be similar to that seen in metazoan origins.
Figures





References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
-
- Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. - PubMed
-
- Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Bielinsky A K, Gerbi S A. Chromosomal ARS1 has a single leading strand start site. Mol Cell. 1999;3:477–486. - PubMed
-
- Bielinsky A K, Gerbi S A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
