Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin
- PMID: 11283265
- PMCID: PMC86916
- DOI: 10.1128/MCB.21.8.2867-2879.2001
Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin
Abstract
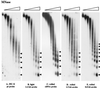
We have used line HS-2 of Drosophila melanogaster, carrying a silenced transgene in the pericentric heterochromatin, to investigate in detail the chromatin structure imposed by this environment. Digestion of the chromatin with micrococcal nuclease (MNase) shows a nucleosome array with extensive long-range order, indicating regular spacing, and with well-defined MNase cleavage fragments, indicating a smaller MNase target in the linker region. The repeating unit is ca. 10 bp larger than that observed for bulk Drosophila chromatin. The silenced transgene shows both a loss of DNase I-hypersensitive sites and decreased sensitivity to DNase I digestion within an array of nucleosomes lacking such sites; within such an array, sensitivity to digestion by MNase is unchanged. The ordered nucleosome array extends across the regulatory region of the transgene, a shift that could explain the loss of transgene expression in heterochromatin. Highly regular nucleosome arrays are observed over several endogenous heterochromatic sequences, indicating that this is a general feature of heterochromatin. However, genes normally active within heterochromatin (rolled and light) do not show this pattern, suggesting that the altered chromatin structure observed is associated with regions that are silent, rather than being a property of the domain as a whole. The results indicate that long-range nucleosomal ordering is linked with the heterochromatic packaging that imposes gene silencing.
Figures







References
-
- Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Position effect variegation at the fission yeast centromere. Cell. 1994;76:157–169. - PubMed
-
- Bellard M, Gannon F, Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harbor Symp Quant Biol. 1978;2:779–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
