Gene expression profiles during the initial phase of salt stress in rice
- PMID: 11283343
- PMCID: PMC135538
- DOI: 10.1105/tpc.13.4.889
Gene expression profiles during the initial phase of salt stress in rice
Abstract
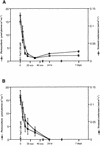
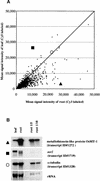
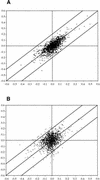
Transcript regulation in response to high salinity was investigated for salt-tolerant rice (var Pokkali) with microarrays including 1728 cDNAs from libraries of salt-stressed roots. NaCl at 150 mM reduced photosynthesis to one tenth of the prestress value within minutes. Hybridizations of RNA to microarray slides probed for changes in transcripts from 15 min to 1 week after salt shock. Beginning 15 min after the shock, Pokkali showed upregulation of transcripts. Approximately 10% of the transcripts in Pokkali were significantly upregulated or downregulated within 1 hr of salt stress. The initial differences between control and stressed plants continued for hours but became less pronounced as the plants adapted over time. The interpretation of an adaptive process was supported by the similar analysis of salinity-sensitive rice (var IR29), in which the immediate response exhibited by Pokkali was delayed and later resulted in downregulation of transcription and death. The upregulated functions observed with Pokkali at different time points during stress adaptation changed over time. Increased protein synthesis and protein turnover were observed at early time points, followed by the induction of known stress-responsive transcripts within hours, and the induction of transcripts for defense-related functions later. After 1 week, the nature of upregulated transcripts (e.g., aquaporins) indicated recovery.
Figures





References
-
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. - PubMed
-
- Barkla, B.J., Vera-Estrella, R., and Pantoja, O. (1999). Towards the production of salt-tolerant crops. Adv. Exp. Med. Biol. 464, 77–89. - PubMed
-
- Basu, S., and Maitra, U. (1988). Salinity results in polyamine accumulation in early rice (Oryza sativa L.) seedlings. Aust. J. Plant Physiol. 15, 777–786.
-
- Berberich, T., and Kusano, T. (1997). Cycloheximide induces a subset of low temperature-inducible genes in maize. Mol. Gen. Genet. 254, 275–283. - PubMed
-
- Boyer, J.S. (1982). Plant productivity and environment. Science 218, 443–448. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

