Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii
- PMID: 11285239
- PMCID: PMC145512
- DOI: 10.1093/emboj/20.7.1765
Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii
Abstract
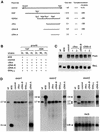
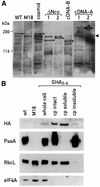
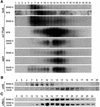
In Chlamydomonas reinhardtii, the psaA mRNA is assembled by a process involving trans-splicing of separate transcripts, encoded at three separate loci of the chloroplast genome. At least 14 nuclear loci and one chloroplast gene, tscA, are needed for this process. We have cloned Raa3, the first nuclear gene implicated in the splicing of intron 1. The predicted sequence of Raa3 consists of 1783 amino acids and shares a small region of homology with pyridoxamine 5'-phosphate oxidases. Raa3 is present in the soluble fraction of the chloroplast and is part of a large 1700 kDa complex, which also contains tscA RNA and the first psaA exon transcript. These partners, in association with other factors, form a chloroplast RNP particle that is required for the splicing of the first intron of psaA and which may be the counterpart of eukaryotic snRNPs involved in nuclear splicing.
Figures


References
-
- Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
-
- Bennoun P. and Delepelaire,P. (1982) Isolation of photosynthesis mutants in Chlamydomonas. In Edelman,M., Hallick,R.B. and Chua,N.H. (eds), Methods of Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp. 25–38.
-
- Cech R.T. (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell, 44, 207–210. - PubMed
-
- Choquet Y., Goldschmidt-Clermont,M., Girard-Bascou,J., Kuck,U., Bennoun,P. and Rochaix,J.D. (1988) Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C.reinhardtii chloroplast. Cell, 52, 903–913. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

