SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs
- PMID: 11285241
- PMCID: PMC145484
- DOI: 10.1093/emboj/20.7.1785
SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs
Abstract
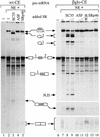
SC35 belongs to the family of SR proteins that regulate alternative splicing in a concentration-dependent manner in vitro and in vivo. We previously reported that SC35 is expressed through alternatively spliced mRNAs with differing 3' untranslated sequences and stabilities. Here, we show that overexpression of SC35 in HeLa cells results in a significant decrease of endogenous SC35 mRNA levels along with changes in the relative abundance of SC35 alternatively spliced mRNAs. Remarkably, SC35 leads to both an exon inclusion and an intron excision in the 3' untranslated region of its mRNAs. In vitro splicing experiments performed with recombinant SR proteins demonstrate that SC35, but not ASF/SF2 or 9G8, specifically activates these alternative splicing events. Interestingly, the resulting mRNA is very unstable and we present evidence that mRNA surveillance is likely to be involved in this instability. SC35 therefore constitutes the first example of a splicing factor that controls its own expression through activation of splicing events leading to expression of unstable mRNA.
Figures













References
-
- Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. - PubMed
-
- Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

