An in vitro evolved precursor tRNA with aminoacylation activity
- PMID: 11285242
- PMCID: PMC145511
- DOI: 10.1093/emboj/20.7.1797
An in vitro evolved precursor tRNA with aminoacylation activity
Abstract
A set of catalysts for aminoacyl-tRNA synthesis is an essential component for translation. The RNA world hypothesis postulates that RNA catalysts could have played this role. Here we show an in vitro evolved precursor tRNA consisting of two domains, a catalytic 5'-leader sequence and an aminoacyl-acceptor tRNA. The 5'-leader sequence domain selectively self-charges phenylalanine on the 3'-terminus of the tRNA domain. This cis-acting ribozyme is susceptible to RNase P RNA, generating the corresponding 5'-leader segment and the mature tRNA. Moreover, the 5'-leader segment is able to aminoacylate the mature tRNA in trans. Mutational studies have revealed that C(74) and C(75) at the tRNA aminoacyl-acceptor end form base pairs with G71 and G70 of the trans-acting ribozyme. Such Watson-Crick base pairing with tRNA has been observed in RNase P RNA and 23S rRNA, suggesting that all three ribozymes use a similar mechanism for the recognition of the aminoacyl-acceptor end. Our demonstrations indicate that catalytic precursor tRNAs could have provided the foundations for the genetic coding system in the proto-translation system.
Figures



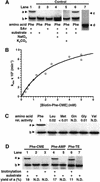
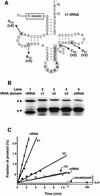
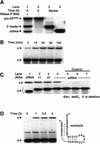
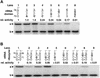
References
-
- Arnez J.G. and Moras,D. (1997) Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci., 22, 211–216. - PubMed
-
- Bartel D.P. and Szostak,J.W. (1993) Isolation of new ribozymes from a large pool of random sequences. Science, 261, 1411–1418. - PubMed
-
- Berg P. (1958) The chemical synthesis of amino acid adenylates. J. Biol. Chem., 253, 608–611. - PubMed
-
- Beuning P.J. and Musier-Forsyth,K. (1999) Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers, 52, 1–28. - PubMed
-
- Busch S., Kirsebom,L.A., Notbohm,H. and Hartmann,R.K. (2000) Differential role of the intermolecular base-pairs G292-C75 and G293-C74 in the reaction catalyzed by Escherichia coli RNase P RNA. J. Mol. Biol., 299, 941–951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

