Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step
- PMID: 11287586
- PMCID: PMC114182
- DOI: 10.1128/JVI.75.9.4376-4385.2001
Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step
Abstract
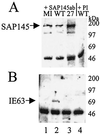
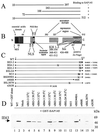
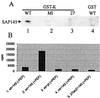
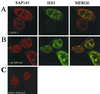
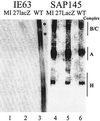
The multifunctional herpes simplex virus type 1 (HSV-1) protein IE63 (ICP27) interacts with the essential pre-mRNA splicing factor, spliceosome-associated protein 145 (SAP145), and in infected cells IE63 and SAP145 colocalize. This interaction was reduced or abrogated completely using extracts from cells infected with IE63 viral mutants, with mutations in IE63 KH and Sm homology domains, which do not exhibit host shutoff or inhibit splicing. In the presence of IE63, splicing in vitro was inhibited prior to the first catalytic step and the B/C complex formed during splicing was shifted up in mobility and reduced in intensity. With the use of splicing extracts, IE63 and SAP145 both comigrated with the B/C complex, suggesting that they interact within this complex to inhibit B/C complex formation or conversion. The inhibition of splicing may facilitate the export of viral or cellular transcripts, possibly via other protein partners of IE63. These data provide important new insights into how IE63 influences pre-mRNA processing during HSV-1 infection.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

