ANX7, a candidate tumor suppressor gene for prostate cancer
- PMID: 11287641
- PMCID: PMC31876
- DOI: 10.1073/pnas.071055798
ANX7, a candidate tumor suppressor gene for prostate cancer
Abstract
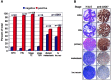
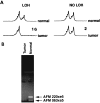
The ANX7 gene is located on human chromosome 10q21, a site long hypothesized to harbor a tumor suppressor gene(s) (TSG) associated with prostate and other cancers. To test whether ANX7 might be a candidate TSG, we examined the ANX7-dependent suppression of human tumor cell growth, stage-specific ANX7 expression in 301 prostate specimens on a prostate tissue microarray, and loss of heterozygosity (LOH) of microsatellite markers at or near the ANX7 locus. Here we report that human tumor cell proliferation and colony formation are markedly reduced when the wild-type ANX7 gene is transfected into two prostate tumor cell lines, LNCaP and DU145. Consistently, analysis of ANX7 protein expression in human prostate tumor microarrays reveals a significantly higher rate of loss of ANX7 expression in metastatic and local recurrences of hormone refractory prostate cancer as compared with primary tumors (P = 0.0001). Using four microsatellite markers at or near the ANX7 locus, and laser capture microdissected tumor cells, 35% of the 20 primary prostate tumors show LOH. The microsatellite marker closest to the ANX7 locus showed the highest rate of LOH, including one homozygous deletion. We conclude that the ANX7 gene exhibits many biological and genetic properties expected of a TSG and may play a role in prostate cancer progression.
Figures


References
-
- Creutz C E, Pazoles C J, Pollard H B. J Biol Chem. 1978;253:2858–2866. - PubMed
-
- Creutz E C, Pazoles C J, Pollard H B. J Biol Chem. 1979;254:553–558. - PubMed
-
- Raynal P, Pollard H B. Biochim Biophys Acta Biomembranes. 1994;1197:63–93. - PubMed
-
- Shirvan A, Srivastava M, Wang M A, Cultraro C, Magendzo K, McBride O W, Pollard H B, Burns A L. Biochemistry. 1994;33:6888–6901. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

