Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach
- PMID: 11287657
- PMCID: PMC31874
- DOI: 10.1073/pnas.081068098
Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach
Abstract
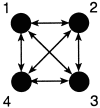
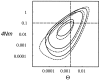
A maximum likelihood estimator based on the coalescent for unequal migration rates and different subpopulation sizes is developed. The method uses a Markov chain Monte Carlo approach to investigate possible genealogies with branch lengths and with migration events. Properties of the new method are shown by using simulated data from a four-population n-island model and a source-sink population model. Our estimation method as coded in migrate is tested against genetree; both programs deliver a very similar likelihood surface. The algorithm converges to the estimates fairly quickly, even when the Markov chain is started from unfavorable parameters. The method was used to estimate gene flow in the Nile valley by using mtDNA data from three human populations.
Figures


References
-
- Weir B S. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

