A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress
- PMID: 11287671
- PMCID: PMC31846
- DOI: 10.1073/pnas.081610398
A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress
Abstract
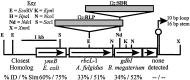
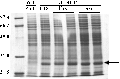
A gene encoding a product with substantial similarity to ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) was identified in the preliminary genome sequence of the green sulfur bacterium Chlorobium tepidum. A highly similar gene was subsequently isolated and sequenced from Chlorobium limicola f.sp. thiosulfatophilum strain Tassajara. Analysis of these amino acid sequences indicated that they lacked several conserved RubisCO active site residues. The Chlorobium RubisCO-like proteins are most closely related to deduced sequences in Bacillus subtilis and Archaeoglobus fulgidus, which also lack some typical RubisCO active site residues. When the C. tepidum gene encoding the RubisCO-like protein was disrupted, the resulting mutant strain displayed a pleiotropic phenotype with defects in photopigment content, photoautotrophic growth and carbon fixation rates, and sulfur metabolism. Most important, the mutant strain showed substantially enhanced accumulation of two oxidative stress proteins. These results indicated that the C. tepidum RubisCO-like protein might be involved in oxidative stress responses and/or sulfur metabolism. This protein might be an evolutional link to bona fide RubisCO and could serve as an important tool to analyze how the RubisCO active site developed.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

