Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay
- PMID: 11290543
- PMCID: PMC1891922
- DOI: 10.1016/S0002-9440(10)64076-X
Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay
Abstract
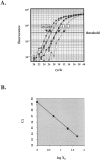
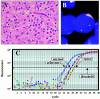
The combined loss of chromosomes 1p and 19q has recently emerged as a genetic predictor of chemosensitivity in anaplastic oligodendrogliomas. Here, we describe a strategy that uses a novel method of real-time quantitative polymerase chain reaction, quantitative microsatellite analysis (QuMA), for the molecular analysis of 1p and 19q loss in oligodendrogliomas and oligoastrocytomas in archival routinely processed paraffin material. QuMA is performed on the ABI 7700 and based on amplifications of microsatellite loci that contain (CA)n repeats where the repeat itself is the target for hybridization by the fluorescently labeled probe. This single probe can therefore be used to determine copy number of microsatellite loci spread throughout the human genome. In genomic DNA prepared from paraffin-embedded brain tumor specimens, QuMA detected combined loss of 1p and 19q in 64% (21 of 32) of oligodendrogliomas and 67% (6 of 9) of oligoastrocytomas. We validate the use of QuMA as a reliable method to detect copy number by showing concordance between QuMA and fluorescence in situ hybridization at 37 of 45 chromosomal arms tested. These results indicate that QuMA is an accurate, high-throughput assay for the detection of copy number at multiple loci; as many as 31 loci of an individual tumor can be analyzed on a 96-well plate in a single 2-hour run. In addition, it has advantages over standard allelic imbalance/loss of heterozygosity assays in that all loci are potentially informative, paired normal tissue is not required, and gain can be distinguished from loss. QuMA may therefore be a powerful molecular tool to expedite the genotypic analysis of human gliomas in a clinical setting for diagnostic/prognostic purposes.
Figures




References
-
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN: Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998, 90:1473-1479 - PubMed
-
- Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB: Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 2000, 18:636-645 - PubMed
-
- Bello MJ, Vaquero J, de Campos JM, Kusak ME, Sarasa JL, Saez-Castresana J, Pestana A, Rey JA: Molecular analysis of chromosome 1 abnormalities in human gliomas reveals frequent loss of 1p in oligodendroglial tumors. Int J Cancer 1994, 57:172-175 - PubMed
-
- Yong WH, Chou D, Ueki K, Harsh GR, IV, von Deimling A, Gusella JF, Mohrenweiser HW, Louis DN: Chromosome 19q deletions in human gliomas overlap telomeric to D19S219 and may target a 425 kb region centromeric to D19S112. J Neuropathol Exp Neurol 1995, 54:622-626 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

