Identification of common binding sites for calmodulin and inositol 1,4,5-trisphosphate receptors on the carboxyl termini of trp channels
- PMID: 11290752
- PMCID: PMC1847329
- DOI: 10.1074/jbc.M102316200
Identification of common binding sites for calmodulin and inositol 1,4,5-trisphosphate receptors on the carboxyl termini of trp channels
Abstract
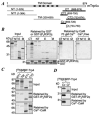
Homologues of Drosophila Trp (transient receptor potential) form plasma membrane channels that mediate Ca(2+) entry following the activation of phospholipase C by cell surface receptors. Among the seven Trp homologous found in mammals, Trp3 has been shown to interact with and respond to IP(3) receptors (IP(3)Rs) for activation. Here we show that Trp4 and other Trp proteins also interact with IP(3)Rs. The IP(3)R-binding domain also interacts with calmodulin (CaM) in a Ca(2+)-dependent manner with affinities ranging from 10 nm for Trp2 to 290 nm for Trp6. In addition, other binding sites for CaM and IP(3)Rs are present in the alpha but not the beta isoform of Trp4. In the presence of Ca(2+), the Trp-IP(3)R interaction is inhibited by CaM. However, a synthetic peptide representing a Trp-binding domain of IP(3)Rs inhibited the binding of CaM to Trp3, -6, and -7 more effectively than that to Trp1, -2, -4, and -5. In inside-out membrane patches, Trp4 is activated strongly by calmidazolium, an antagonist of CaM, and a high (50 microm) but not a low (5 microm) concentration of the Trp-binding peptide of the IP(3)R. Our data support the view that both CaM and IP(3)Rs play important roles in controlling the gating of Trp-based channels. However, the sensitivity and responses to CaM and IP(3)Rs differ for each Trp.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

