BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition
- PMID: 11292725
- PMCID: PMC98261
- DOI: 10.1128/IAI.69.5.3067-3072.2001
BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition
Abstract
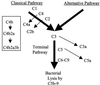
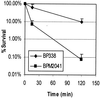
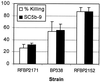
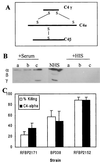
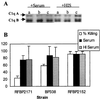
Bordetella pertussis produces a 73-kDa protein, BrkA (Bordetella resistance to killing), which inhibits the bactericidal activity of complement. In this study we characterized the step in the complement cascade where BrkA acts, using three strains: a wild-type strain, a strain containing an insertional disruption of brkA, and a strain containing two copies of the brkA locus. Following incubation with 10% human serum, killing was greatest for the BrkA mutant, followed by that for the wild-type strain, while the strain with two copies of brkA was the most resistant. Complement activation was monitored by enzyme-linked immunosorbent assay (ELISA) or Western blotting. ELISAs for SC5b-9, the soluble membrane attack complex, showed that production of SC5b-9 was greatest with the brkA mutant, less with the wild type, and least with the strain containing two copies of brkA. Deposition of complement proteins on the bacteria was monitored by Western blotting. A decrease in deposition on the bacteria of C4, C3, and C9 corresponded with decreased complement sensitivity. Deposition of C1, however, was not affected by the presence of BrkA. These studies show that BrkA inhibits the classical pathway of complement activation and prevents accumulation of deposited C4.
Figures



References
-
- Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. - PubMed
-
- Bock S C, Skriver K, Nielsen E, Thogersen H C, Wiman B, Donaldson V H, Eddy R L, Marrinan J, Radziejewska E, Huber R, et al. Human C1 inhibitor: primary structure, cDNA cloning, and chromosomal localization. Biochemistry. 1986;25:4292–4301. - PubMed
-
- Brzezinski R M, Boutelet-Bochan H, Person R E, Fantel A G, Juchau M R. Catalytic activity and quantitation of cytochrome P-450 2E1 in prenatal human brain. J Pharmacol Exp Ther. 1999;289:1648–1653. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

