Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B
- PMID: 11292803
- PMCID: PMC99500
- DOI: 10.1128/JB.183.9.2834-2841.2001
Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B
Abstract
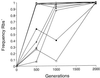
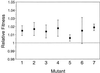
Twelve populations of Escherichia coli B all lost D-ribose catabolic function during 2,000 generations of evolution in glucose minimal medium. We sought to identify the population genetic processes and molecular genetic events that caused these rapid and parallel losses. Seven independent Rbs(-) mutants were isolated, and their competitive fitnesses were measured relative to that of their Rbs(+) progenitor. These Rbs(-) mutants were all about 1 to 2% more fit than the progenitor. A fluctuation test revealed an unusually high rate, about 5 x 10(-5) per cell generation, of mutation from Rbs(+) to Rbs(-), which contributed to rapid fixation. At the molecular level, the loss of ribose catabolic function involved the deletion of part or all of the ribose operon (rbs genes). The physical extent of the deletion varied between mutants, but each deletion was associated with an IS150 element located immediately upstream of the rbs operon. The deletions apparently involved transposition into various locations within the rbs operon; recombination between the new IS150 copy and the one upstream of the rbs operon then led to the deletion of the intervening sequence. To confirm that the beneficial fitness effect was caused by deletion of the rbs operon (and not some undetected mutation elsewhere), we used P1 transduction to restore the functional rbs operon to two Rbs(-) mutants, and we constructed another Rbs(-) strain by gene replacement with a deletion not involving IS150. All three of these new constructs confirmed that Rbs(-) mutants have a competitive advantage relative to their Rbs(+) counterparts in glucose minimal medium. The rapid and parallel evolutionary losses of ribose catabolic function thus involved both (i) an unusually high mutation rate, such that Rbs(-) mutants appeared repeatedly in all populations, and (ii) a selective advantage in glucose minimal medium that drove these mutants to fixation.
Figures

References
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
-
- Blot M. Transposable elements and adaptation of host bacteria. Genetica. 1994;93:5–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

