Function-based selection and characterization of base-pair polymorphisms in a promoter of Escherichia coli RNA polymerase-sigma(70)
- PMID: 11292807
- PMCID: PMC99504
- DOI: 10.1128/JB.183.9.2866-2873.2001
Function-based selection and characterization of base-pair polymorphisms in a promoter of Escherichia coli RNA polymerase-sigma(70)
Abstract
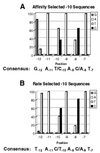
We performed two sets of in vitro selections to dissect the role of the -10 base sequence in determining the rate and efficiency with which Escherichia coli RNA polymerase-sigma(70) forms stable complexes with a promoter. We identified sequences that (i) rapidly form heparin-resistant complexes with RNA polymerase or (ii) form heparin-resistant complexes at very low RNA polymerase concentrations. The sequences selected under the two conditions differ from each other and from the consensus -10 sequence. The selected promoters have the expected enhanced binding and kinetic properties and are functionally better than the consensus promoter sequence in directing RNA synthesis in vitro. Detailed analysis of the selected promoter functions shows that each step in this multistep pathway may have different sequence requirements, meaning that the sequence of a strong promoter does not contain the optimal sequence for each step but instead is a compromise sequence that allows all steps to proceed with minimal constraint.
Figures


Similar articles
-
Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes.J Mol Biol. 2001 Jun 8;309(3):561-72. doi: 10.1006/jmbi.2001.4690. J Mol Biol. 2001. PMID: 11397080
-
Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex.J Mol Biol. 2000 Jun 23;299(5):1217-30. doi: 10.1006/jmbi.2000.3808. J Mol Biol. 2000. PMID: 10873447
-
The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli.PLoS One. 2014 Mar 6;9(3):e90447. doi: 10.1371/journal.pone.0090447. eCollection 2014. PLoS One. 2014. PMID: 24603758 Free PMC article.
-
Protein-nucleic acid interactions in transcription: a molecular analysis.Annu Rev Biochem. 1984;53:389-446. doi: 10.1146/annurev.bi.53.070184.002133. Annu Rev Biochem. 1984. PMID: 6206781 Review. No abstract available.
-
Role of the RNA polymerase sigma subunit in transcription initiation.Res Microbiol. 2002 Nov;153(9):557-62. doi: 10.1016/s0923-2508(02)01368-2. Res Microbiol. 2002. PMID: 12455702 Review.
Cited by
-
Efficient bacterial transcription of DNA nanocircle vectors with optimized single-stranded promoters.Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):54-9. doi: 10.1073/pnas.012589099. Epub 2001 Dec 18. Proc Natl Acad Sci U S A. 2002. PMID: 11752404 Free PMC article.
-
Alteration of the -35 and -10 sequences and deletion the upstream sequence of the -35 region of the promoter A1 of the phage T7 in dsDNA confirm the contribution of non-specific interactions with E. coli RNA polymerase to the transcription initiation process.Front Mol Biosci. 2024 Jan 8;10:1335409. doi: 10.3389/fmolb.2023.1335409. eCollection 2023. Front Mol Biosci. 2024. PMID: 38259683 Free PMC article.
-
Strategies for enhancing bioluminescent bacterial sensor performance by promoter region manipulation.Bioeng Bugs. 2010 Mar-Apr;1(2):151-3. doi: 10.4161/bbug.1.2.11104. Epub 2010 Jan 14. Bioeng Bugs. 2010. PMID: 21326942 Free PMC article.
-
Strategies for enhancing bioluminescent bacterial sensor performance by promoter region manipulation.Microb Biotechnol. 2010 May;3(3):300-10. doi: 10.1111/j.1751-7915.2009.00149.x. Epub 2009 Aug 26. Microb Biotechnol. 2010. PMID: 21255329 Free PMC article.
-
The transition from transcriptional initiation to elongation.Curr Opin Genet Dev. 2008 Apr;18(2):130-6. doi: 10.1016/j.gde.2007.12.008. Epub 2008 Feb 20. Curr Opin Genet Dev. 2008. PMID: 18282700 Free PMC article. Review.
References
-
- Buc H, McClure W R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. - PubMed
-
- Bushman F D. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J Mol Biol. 1993;230:28–40. - PubMed
-
- Craig M L, Suh W C, Record M T., Jr HO⋅ and DNase I probing of E sigma 70 RNA polymerase-lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

