Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy
- PMID: 11296244
- PMCID: PMC33154
- DOI: 10.1073/pnas.071372498
Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy
Abstract
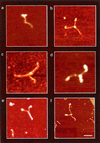
The Escherichia coli protein DbpA is unique in its subclass of DEAD box RNA helicases, because it possesses ATPase-specific activity toward the peptidyl transferase center in 23S rRNA. Although its remarkable ATPase activity had been well defined toward various substrates, its RNA helicase activity remained to be characterized. Herein, we show by using biochemical assays and atomic force microscopy that DbpA exhibits ATP-stimulated unwinding activity of RNA duplex regardless of its primary sequence. This work presents an attempt to investigate the action of DEAD box proteins by a single-molecule visualization methodology. Our atomic force microscopy images enabled us to observe directly the unwinding reaction of a DEAD box helicase on long stretches of double-stranded RNA. Specifically, we could differentiate between the binding of DbpA to RNA in the absence of ATP and the formation of a Y-shaped intermediate after its progression through double-stranded RNA in the presence of ATP. Recent studies have questioned the designation of DbpA, in particular, and DEAD box proteins in general as RNA helicases. However, accumulated evidence and the results reported herein suggest that these proteins are indeed helicases that resemble in many aspects the DNA helicases.
Figures




References
-
- Linder P, Lasko P F, Ashburner M, Leroy P, Nielsen P J, Nishi K, Schnier J, Slonimski P P. Nature (London) 1989;337:121–122. - PubMed
-
- de la Cruz J, Kressler D, Linder P. Trends Biochem Sci. 1999;24:192–198. - PubMed
-
- Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Nature (London) 1988;333:22. (lett.). - PubMed
-
- Egelman E H. J Struct Biol. 1998;124:123–128. - PubMed
-
- Luking A, Stahl U, Schmidt U. Crit Rev Biochem Mol Biol. 1998;33:259–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

