Complete genome sequence of an M1 strain of Streptococcus pyogenes
- PMID: 11296296
- PMCID: PMC31890
- DOI: 10.1073/pnas.071559398
Complete genome sequence of an M1 strain of Streptococcus pyogenes
Abstract
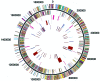
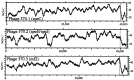
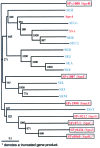
The 1,852,442-bp sequence of an M1 strain of Streptococcus pyogenes, a Gram-positive pathogen, has been determined and contains 1,752 predicted protein-encoding genes. Approximately one-third of these genes have no identifiable function, with the remainder falling into previously characterized categories of known microbial function. Consistent with the observation that S. pyogenes is responsible for a wider variety of human disease than any other bacterial species, more than 40 putative virulence-associated genes have been identified. Additional genes have been identified that encode proteins likely associated with microbial "molecular mimicry" of host characteristics and involved in rheumatic fever or acute glomerulonephritis. The complete or partial sequence of four different bacteriophage genomes is also present, with each containing genes for one or more previously undiscovered superantigen-like proteins. These prophage-associated genes encode at least six potential virulence factors, emphasizing the importance of bacteriophages in horizontal gene transfer and a possible mechanism for generating new strains with increased pathogenic potential.
Figures
References
-
- Stevens D L, Tanner M H, Winship J, Swarts R, Ries K M, Schlievert P M, Kaplan E. N Engl J Med. 1989;321:1–7. - PubMed
-
- Veasy L G, Wiedmeier S E, Orsmond G S. N Engl J Med. 1987;316:421–427. - PubMed
-
- Bodenteich A, Chissoe S, Wang Y F, Roe B A. In: Automated DNA Sequencing and Analysis Techniques. Ventor C, editor. London: Academic; 1993.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

