The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes
- PMID: 11297506
- PMCID: PMC312663
- DOI: 10.1101/gad.876201
The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes
Abstract
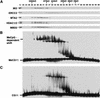
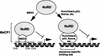
Histone deacetylation plays an important role in methylated DNA silencing. Recent studies indicated that the methyl-CpG-binding protein, MBD2, is a component of the MeCP1 histone deacetylase complex. Interestingly, MBD2 is able to recruit the nucleosome remodeling and histone deacetylase, NuRD, to methylated DNA in vitro. To understand the relationship between the MeCP1 complex and the NuRD complex, we purified the MeCP1 complex to homogeneity and found that it contains 10 major polypeptides including MBD2 and all of the known NuRD components. Functional analysis of the purified MeCP1 complex revealed that it preferentially binds, remodels, and deacetylates methylated nucleosomes. Thus, our study defines the MeCP1 complex, and provides biochemical evidence linking nucleosome remodeling and histone deacetylation to methylated gene silencing.
Figures

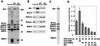

References
-
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. - PubMed
-
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. - PubMed
-
- Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. - PubMed
-
- Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:504–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
