ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis
- PMID: 11297513
- PMCID: PMC312669
- DOI: 10.1101/gad.866201
ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis
Abstract
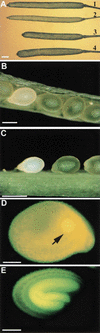
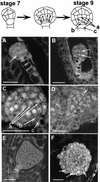
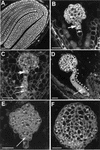
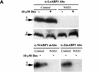
To directly address the function of a putative auxin receptor designated ABP1, a reverse genetic approach was taken to identify and characterize ABP1 mutant alleles in Arabidopsis. A homozygous null mutation in ABP1 confers embryo lethality. Null mutant embryos develop normally until the early stages of the globular embryo but are unable to make the transition to a bilaterally symmetrical structure because cells fail to elongate. Cell division was also aberrant both in the suspensor and embryo proper. Antisense suppression of ABP1 in tobacco cells causes slow proliferation and eliminates auxin-induced cell elongation and reduces cell division. The complete lack of auxin-inducible elongation in individual cells confirms the results observed in embryos, indicates a cell autonomous function, and, taken together with biochemical evidence that ABP1 binds auxins, suggests that ABP1 mediates auxin-induced cell elongation and, directly or indirectly, cell division.
Figures





Similar articles
-
Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco.Plant Cell. 2008 Oct;20(10):2746-62. doi: 10.1105/tpc.108.059048. Epub 2008 Oct 24. Plant Cell. 2008. PMID: 18952781 Free PMC article.
-
Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2275-80. doi: 10.1073/pnas.1500365112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646447 Free PMC article.
-
Complementation of the embryo-lethal T-DNA insertion mutant of AUXIN-BINDING-PROTEIN 1 (ABP1) with abp1 point mutated versions reveals crosstalk of ABP1 and phytochromes.J Exp Bot. 2015 Jan;66(1):403-18. doi: 10.1093/jxb/eru433. Epub 2014 Nov 11. J Exp Bot. 2015. PMID: 25392478 Free PMC article.
-
Is ABP1 an auxin receptor yet?Mol Plant. 2011 Jul;4(4):635-40. doi: 10.1093/mp/ssr050. Epub 2011 Jun 29. Mol Plant. 2011. PMID: 21715649 Free PMC article. Review.
-
AUXIN BINDING PROTEIN 1: functional and evolutionary aspects.Trends Plant Sci. 2010 Aug;15(8):436-46. doi: 10.1016/j.tplants.2010.05.001. Epub 2010 Jun 3. Trends Plant Sci. 2010. PMID: 20605513 Review.
Cited by
-
No Time for Transcription-Rapid Auxin Responses in Plants.Cold Spring Harb Perspect Biol. 2021 Aug 2;13(8):a039891. doi: 10.1101/cshperspect.a039891. Cold Spring Harb Perspect Biol. 2021. PMID: 33648988 Free PMC article. Review.
-
Sequence evaluation of four specific cDNA libraries for developmental genomics of sunflower.Mol Genet Genomics. 2004 Apr;271(3):367-75. doi: 10.1007/s00438-004-0989-5. Epub 2004 Mar 9. Mol Genet Genomics. 2004. PMID: 15007731
-
The auxin-induced K(+) channel gene Zmk1 in maize functions in coleoptile growth and is required for embryo development.Plant Mol Biol. 2006 Jul;61(4-5):757-68. doi: 10.1007/s11103-006-0047-2. Plant Mol Biol. 2006. PMID: 16897490
-
The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes.Plant Cell. 2003 Feb;15(2):393-409. doi: 10.1105/tpc.006148. Plant Cell. 2003. PMID: 12566580 Free PMC article.
-
New insights in shoot apical meristem morphogenesis: Isotropy comes into play.Plant Signal Behav. 2015;10(11):e1000150. doi: 10.1080/15592324.2014.1000150. Plant Signal Behav. 2015. PMID: 26337646 Free PMC article.
References
-
- Barbier-Brygoo H, Ephritikhine G, Klambt D, Maurel C, Palme K, Schell J, Guern J. Perception of the auxin signal at the plasma membrane of tobacco mesophyll protoplasts. Plant J. 1991;1:83–93. - PubMed
-
- Batt S, Wilkens MB, Venis MA. Auxin binding to corn coleoptile membranes: Kinetics and specificity. Planta. 1976;130:7–13. - PubMed
-
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. - PubMed
-
- Birnbaumer L, Pohl SL, Kaumann AJ. Receptors and acceptors: A necessary distinction in hormone binding studies. In: Greengard P, Robison GG, editors. Advances in cyclic nucleotide research. New York: North-Holland; 1974. pp. 239–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
