Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families
- PMID: 11299383
- PMCID: PMC88859
- DOI: 10.1104/pp.125.4.2040
Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families
Abstract
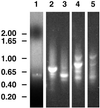
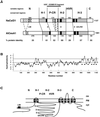
The walls deposited by growing pollen tubes contain two types of beta-glucan, the (1,3)-beta-glucan callose and the (1,4)-beta-glucan cellulose, as well as various alpha-linked pectic polysaccharides. Pollen tubes of Nicotiana alata Link et Otto, an ornamental tobacco, were therefore used to identify genes potentially encoding catalytic subunits of the callose synthase and cellulose synthase enzymes. Reverse transcriptase-polymerase chain reactions (RT-PCR) with pollen-tube RNA and primers designed to conserved regions of bacterial and plant cellulose synthase (CesA) genes amplified a fragment that corresponded to an abundantly expressed cellulose-synthase-like gene named NaCslD1. A fragment from a true CesA gene (NaCesA1) was also amplified, but corresponding cDNAs could not be identified in a pollen-tube library, consistent with the very low level of expression of the NaCesA1 gene. RT-PCR with pollen-tube RNA and primers designed to regions conserved between the fungal FKS genes [that encode (1,3)-beta-glucan synthases] and their presumed plant homologs (the Gsl or glucan-synthase-like genes) amplified a fragment that corresponded to an abundantly expressed gene named NaGsl1. A second Gsl gene detected by RT-PCR (NaGsl2) was expressed at low levels in immature floral organs. The structure of full-length cDNAs of NaCslD1, NaCesA1, and NaGsl1 are presented. Both NaCslD1 and NaGsl1 are predominantly expressed in the male gametophyte (developing and mature pollen and growing pollen tubes), and we propose that they encode the catalytic subunits of two beta-glucan synthases involved in pollen-tube wall synthesis. Different beta-glucans deposited in one cell type may therefore be synthesized by enzymes from different gene families.
Figures

References
-
- Antelo L, Cosio EG, Hertkorn N, Ebel J. Partial purification of a GTP-insensitive (1,3)-β-glucan synthase from Phytophthora sojae. FEBS Lett. 1998;433:191–195. - PubMed
-
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. - PubMed
-
- Bacic A, Harris PJ, Stone BA. Structure and function of plant cell walls. In: Priess J, editor. The Biochemistry of Plants. New York: Academic Press; 1988. pp. 297–371.
-
- Becker M, Vincent C, Reid JSG. Biosynthesis of (1,3)(1,4)-β-glucan and (1,3)-β-glucan in barley (Hordeum vulgare L): properties of the membrane-bound glucan synthases. Planta. 1995;195:331–338. - PubMed
-
- Billongrand C, Marais MF, Joseleau JP, Girard V, Gay L, Fevre M. A novel 1,3-β-glucan synthase from the oomycete Saprolegnia monoica. Microbiology. 1997;143:3175–3187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

