The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families
- PMID: 11304550
- PMCID: PMC2193401
- DOI: 10.1084/jem.193.8.893
The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families
Abstract
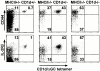
To define the phenotype and T cell receptor (TCR) repertoire of CD1d-dependent T cells, we compared the populations of T cells that persisted in major histocompatibility complex (MHC)-deficient mice, which lack mainstream T cells, with those from MHC/CD1d doubly deficient mice, which lack both mainstream and CD1d-dependent T cells. Surprisingly, up to 80% of the CD1d-dependent T cells were stained by tetramers of CD1d/alpha-galactosylceramide, which specifically identify the previously described CD1d autoreactive Valpha14-Jalpha18/Vbeta8 natural killer (NK) T cells. Furthermore, zooming in on the CD1d-dependent non-Valpha14 T cells, we found that, like Valpha14 NK T cells, they mainly expressed recurrent, CD1d autoreactive TCR families and had a natural memory phenotype. Thus, CD1d-restricted T cells differ profoundly from MHC-peptide-specific T cells by their predominant use of autoreactive and semiinvariant, rather than naive and diverse, TCRs. They more closely resemble other lineages of innate lymphocytes such as B-1 B cells, gammadelta T cells, and NK cells, which express invariant or semiinvariant autoreactive receptors. Finally, we demonstrate that the MHC-restricted TCR repertoire is essentially non-cross-reactive to CD1d. Altogether, these findings imply that lipid recognition by CD1d-restricted T cells may have largely evolved as an innate rather than an adaptive arm of the mouse immune system.
Figures
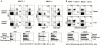


References
-
- Kasahara M., Nakaya J., Satta Y., Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. - PubMed
-
- Calabi F., Jarvis J.M., Martin L.H., Milstein C. Two classes of CD1 genes. Eur. J. Immunol. 1989;19:285–292. - PubMed
-
- Porcelli S.A., Modlin R.L. The CD1 systemantigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 1999;17:297–329. - PubMed
-
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E. CD1d-restricted and TCR-mediated activation of Vα14 NK T cells by glycosylceramides. Science. 1997;278:1626–1629. - PubMed
-
- Porcelli S., Morita C.T., Brenner M.B. CD1β restricts the response of human CD4-8− T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

