A basis for a visual language for describing, archiving and analyzing functional models of complex biological systems
- PMID: 11305940
- PMCID: PMC31481
- DOI: 10.1186/gb-2001-2-4-research0012
A basis for a visual language for describing, archiving and analyzing functional models of complex biological systems
Abstract
Background: We propose that a computerized, internet-based graphical description language for systems biology will be essential for describing, archiving and analyzing complex problems of biological function in health and disease.
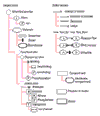
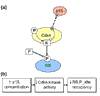
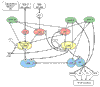
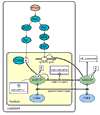
Results: We outline here a conceptual basis for designing such a language and describe BioD, a prototype language that we have used to explore the utility and feasibility of this approach to functional biology. Using example models, we demonstrate that a rather limited lexicon of icons and arrows suffices to describe complex cell-biological systems as discrete models that can be posted and linked on the internet.
Conclusions: Given available computer and internet technology, BioD may be implemented as an extensible, multidisciplinary language that can be used to archive functional systems knowledge and be extended to support both qualitative and quantitative functional analysis.
Figures


References
-
- Karp PD. Metabolic databases. Trends Biochem Sci. 1998;23:114–116. - PubMed
-
- From sequence to function. An introduction to the KEGG project http://kegg.genome.ad.jp/kegg/kegg2.html
-
- McAdams HH, Shapiro L. Circuit simulation of genetic networks. Science. 1995;269:650–656. - PubMed
-
- Takai-Igarashi T, Nadaoka Y, Kaminuma T. A database for cell signaling networks. J Comput Biol. 1998;5:747–754. - PubMed
-
- Transpath home page http://193.175.244.148/index.html
MeSH terms
LinkOut - more resources
Full Text Sources

