Dynamics of mobile element activity in chalcone synthase loci in the common morning glory (Ipomoea purpurea)
- PMID: 11309503
- PMCID: PMC33167
- DOI: 10.1073/pnas.091095498
Dynamics of mobile element activity in chalcone synthase loci in the common morning glory (Ipomoea purpurea)
Abstract
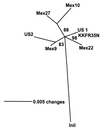
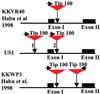
Mobile element dynamics in seven alleles of the chalcone synthase D locus (CHS-D) of the common morning glory (Ipomoea purpurea) are analyzed in the context of synonymous nucleotide sequence distances for CHS-D exons. By using a nucleotide sequence of CHS-D from the sister species Ipomoea nil (Japanese morning glory [Johzuka-Hisatomi, Y., Hoshino, A., Mori, T., Habu, Y. & Iida, S. (1999) Genes Genet. Syst. 74, 141-147], it is also possible to determine the relative frequency of insertion and loss of elements within the CHS-D locus between these two species. At least four different types of transposable elements exist upstream of the coding region, or within the single intron of the CHS-D locus in I. purpurea. There are three distinct families of miniature inverted-repeat transposable elements (MITES), and some recent transpositions of Activator/Dissociation (Ac/Ds)-like elements (Tip100), of some short interspersed repetitive elements (SINEs), and of an insertion sequence (InsIpCHSD) found in the neighborhood of this locus. The data provide no compelling evidence of the transposition of the mites since the separation of I. nil and I. purpurea roughly 8 million years ago. Finally, it is shown that the number and frequency of mobile elements are highly heterogeneous among different duplicate CHS loci, suggesting that the dynamics observed at CHS-D are locus-specific.
Figures


Similar articles
-
The molecular basis of incomplete dominance at the A locus of CHS-D in the common morning glory, Ipomoea purpurea.J Plant Res. 2011 Mar;124(2):299-304. doi: 10.1007/s10265-010-0369-7. Epub 2010 Aug 3. J Plant Res. 2011. PMID: 20680382
-
Molecular evolution of the chalcone synthase multigene family in the morning glory genome.Plant Mol Biol. 2000 Jan;42(1):79-92. Plant Mol Biol. 2000. PMID: 10688131 Review.
-
The transposon Tip100 from the common morning glory is an autonomous element that can transpose in tobacco plants.Mol Genet Genomics. 2002 Jan;266(5):732-9. doi: 10.1007/s00438-001-0603-z. Epub 2001 Nov 8. Mol Genet Genomics. 2002. PMID: 11810246
-
Evolution of the chalcone synthase gene family in the genus Ipomoea.Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3338-42. doi: 10.1073/pnas.92.8.3338. Proc Natl Acad Sci U S A. 1995. PMID: 7724563 Free PMC article.
-
Floricultural traits and transposable elements in the Japanese and common morning glories.Ann N Y Acad Sci. 1999 May 18;870:265-74. doi: 10.1111/j.1749-6632.1999.tb08887.x. Ann N Y Acad Sci. 1999. PMID: 10415489 Review.
Cited by
-
The molecular basis of incomplete dominance at the A locus of CHS-D in the common morning glory, Ipomoea purpurea.J Plant Res. 2011 Mar;124(2):299-304. doi: 10.1007/s10265-010-0369-7. Epub 2010 Aug 3. J Plant Res. 2011. PMID: 20680382
-
Transposable elements are enriched within or in close proximity to xenobiotic-metabolizing cytochrome P450 genes.BMC Evol Biol. 2007 Mar 23;7:46. doi: 10.1186/1471-2148-7-46. BMC Evol Biol. 2007. PMID: 17381843 Free PMC article.
-
Multiple MYB Activators and Repressors Collaboratively Regulate the Juvenile Red Fading in Leaves of Sweetpotato.Front Plant Sci. 2020 Jun 25;11:941. doi: 10.3389/fpls.2020.00941. eCollection 2020. Front Plant Sci. 2020. PMID: 32670334 Free PMC article.
-
Genome-wide identification, evolution, and expression and metabolic regulation of the maize CHS gene family under abiotic stress.BMC Genomics. 2025 Jul 1;26(1):581. doi: 10.1186/s12864-025-11761-0. BMC Genomics. 2025. PMID: 40597569 Free PMC article.
References
-
- Clegg M T. In: Evolutionary Processes and Theory. Waser S P, editor. Dordrecht, The Netherlands: Kluwer; 1999. pp. 55–64.
-
- Bennetzen J L. Plant Mol Biol. 2000;42:251–269. - PubMed
-
- Clegg M T. J Hered. 1997;88:1–7. - PubMed
-
- Iida S, Hoshino A, Johzuka-Hisatomi Y, Habu Y, Inagaki Y. Ann NY Acad Sci. 1999;870:265–274. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

