Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p
- PMID: 11313466
- PMCID: PMC100262
- DOI: 10.1128/MCB.21.10.3405-3415.2001
Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p
Abstract
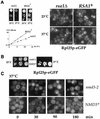
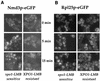
Nuclear export of ribosomes requires a subset of nucleoporins and the Ran system, but specific transport factors have not been identified. Using a large subunit reporter (Rpl25p-eGFP), we have isolated several temperature-sensitive ribosomal export (rix) mutants. One of these corresponds to the ribosomal protein Rpl10p, which interacts directly with Nmd3p, a conserved and essential protein associated with 60S subunits. We find that thermosensitive nmd3 mutants are impaired in large subunit export. Strikingly, Nmd3p shuttles between the nucleus and cytoplasm and is exported by the nuclear export receptor Xpo1p. Moreover, we show that export of 60S subunits is Xpo1p dependent. We conclude that nuclear export of 60S subunits requires the nuclear export sequence-containing nonribosomal protein Nmd3p, which directly binds to the large subunit protein Rpl10p.
Figures



References
-
- Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Dick F A, Karamanou S, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex, codes for a 60 S ribosomal subunit protein. J Biol Chem. 1997;272:13372–13379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
